Abstract
Background
The increase in detections of Plasmodium vivax infection in Duffy-negative individuals in Africa has challenged the dogma establishing the unique P. vivax Duffy Binding Protein-Duffy antigen receptor for chemokines (PvDBP-DARC) pathway used by P. vivax merozoites to invade reticulocytes. Information on the impact of Duffy antigen polymorphisms on the epidemiology of P. vivax malaria remains elusive. The objective of this study was to determine the distribution of asexual parasitaemia of P. vivax according to the Duffy antigen polymorphisms in Ethiopia.
Methods
DNA was extracted from dried blood spots (DBS) collected from prospectively recruited 138 P. vivax-infected patients from health centres. The identification and estimation of P. vivax asexual parasitaemia were performed by microscopic examination and quantitative real-time polymerase chain reaction (PCR). Duffy genotyping was conducted by DNA sequencing in a total of 138 P.vivax infected samples.
Results
The proportion of Duffy-negatives (FY*BES/FY*BES) in P. vivax infected patients was 2.9% (4/138). Duffy genotype FY*B/FY*BES (48.6%) was the most common, followed by FY*A/FY*BES genotype (25.4%). In one patient, the FY*02 W.01/FY*02 N.01 genotype conferring a weak expression of the Fyb antigen was observed. All P.vivax infected Duffy-negative patients showed low asexual parasitaemia (≤ 110 parasites/µL). The median P. vivax parasitaemia in Duffy-negative patients (53 parasites/µL) was significantly lower than those found in homozygous and heterozygous individuals (P < 0.0001).
Conclusion
Plasmodium vivax in Duffy-negative patients shows invariably low asexual parasitaemia. This finding suggests that the pathway used by P. vivax to invade Duffy-negative reticulocytes is much less efficient than that used in Duffy-positives. Moreover, the low asexual parasitaemia observed in Duffy-negative individuals could constitute an ‘undetected silent reservoir', thus likely delaying the elimination of vivax malaria in Ethiopia.
Similar content being viewed by others
Background
In 2020, the World Health Organization (WHO) estimated that malaria contributed to cause 241 million cases and 627,000 deaths [1]. Most malaria cases were caused by Plasmodium falciparum, while recently the prevalence and severity of Plasmodium vivax malaria cases has shown a sharp increment [2]. Plasmodium vivax is a Plasmodium species capable to survive even in temperate climates, which explains its wider range of geographical distribution than other species [3]. In East Shewa, Ethiopia, P. vivax has been responsible for 54–71.4% of malaria cases [4, 5].
Unlike P. falciparum, the invasion of reticulocytes by P. vivax requires the dual interactions between two parasite ligands (P. vivax Duffy Binding Protein, PvDBP and P. vivax reticulocyte Binding Protein 2b, PvRBP2b) and two human receptors (Duffy antigen/receptor for chemokines, DARC/CD234 and transferrin receptor 1, TfR1/CD71) [6,7,8].
The Duffy antigen has Fya and Fyb variants differing from each other by a single nucleotide polymorphism of replacing Glycine to Aspartic acid at codon 42. Encoded by the FY gene (ACKR1 located on chromosome 1q23.2), the Duffy glycoprotein has FY*01 (FY*A) and FY*02 (FY*B) codominant alleles. In other words, if the FY*A is inherited from one parent, the FY*B allele will be inherited from the other. Both gene products, Fya and Fyb antigens are expressed on the red blood cells (RBCs). There are four main Duffy phenotypes: Fy(a + b +), Fy(a + b −), Fy(a − b +), and Fy(a − b −). The last one gives rise to Duffy-negative phenotypes [9, 10] and was thought to confer innate resistance to blood-stage infection with P. vivax and clinical malaria [11]. Recently, however, the widespread detection of P. vivax across individuals lacking Duffy antigen in many African countries [9, 12,13,14,15,16,17] including Ethiopia [13, 18, 19] further complicates the current understanding of vivax malaria epidemiology in the region.
It has been postulated that hidden transmission could be occurring in Duffy-negative populations in sub-Saharan Africa. A plausible explanation that could support this notion is the observation of Duffy-independent P. vivax blood-stage infection across the Duffy-negative population of Africa [20]. Most of the previous reports have given more emphasis on P. vivax DNA detection in Duffy-negative individuals by polymerase chain reaction (PCR) without parasite density estimation. It has been suggested that the expression of Fya and Fyb antigens on the reticulocytes influenced asexual P. vivax parasitaemia, thereby modulating susceptibility to clinical vivax malaria [21]. For instance, in Papua New Guinea, it has been shown that Duffy heterozygous (FY*A/FY*BES) individuals had lower P. vivax parasitaemia than those individuals with homozygous (FY*A/FY*A) [22]. To date, the association of clinical P. vivax with Duffy antigen is contradicting. For instance, genotypes FY*A/FY*A, as compared to FY*B/FY*B, were reported in the increasing susceptibility of individuals and the severity of P. vivax infection [23]. In contrary, some studies have shown that FY*B/FY*B genotypes were highly susceptible to P. vivax malaria [21, 24,25,26].
In Ethiopia, data regarding the epidemiological features of vivax malaria in relation with duffy blood group are scarce [13, 18, 19], while an admixture of people with varying Duffy antigen polymorphism co-exists and P. vivax transmission is frequent. This provides the opportunity to study distribution and effect of Duffy antigen polymorphisms on the asexual P. vivax parasitaemia. This study was, therefore, initiated to explore this question in the admixed populations of Duffy-positive and Duffy-negative individuals in East Shewa, Ethiopia.
Methods
Study area and population
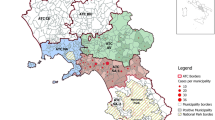
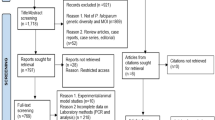
The study was conducted in Adama City Administration, and its surroundings located 100 km southeast of Addis Ababa, the capital city of Ethiopia from October 2019 up to January 2021 (Fig. 1). Adama City Administration is presented on 1623 m above sea level, 20.5 °C average annual temperature and 808 mm annual rainfalls. Plasmodium vivax is reported as the predominant malaria-causing species [4, 5] in East Shewa. A total of 151 individuals were prospectively recruited from all health centres. The study populations were those patients with fever seeking health care in public health institutions at Adama City Administratin (malaria diagnostic centre, and 7 health centres), and surroundings (Awash Melkasa, Koka, Metehara, Mojo and Welenchity health centres) in East Shewa Zone. All patients known to have uncomplicated P. vivax malaria by microscopy were included when possible. Those with P. falciparum and a recent history of antimalarial drugs were excluded from the current study.
Blood sample collection
Clinical P. vivax blood samples were collected from patients seeking malaria diagnosis and treatment at health centres. Finger-prick blood samples were collected to determine Plasmodium infection. Both thin and thick blood films were prepared in a single slide and labeled with the identification number of participants and date of preparation. The thin smear was fixed with methanol, and both thick and thin blood smears were stained with Giemsa according to the protocol [27]. Each slide was examined independently by two experienced microscopists who were blinded to each other’s result under a microscope with 100 × oil immersion. Slides with discordant results were read by a third reader blindly, and the majority results were taken as final. If a P. vivax infection was confirmed by microscopy examination, two to four spots of blood, near to ~ 60 μl each spot, were collected on Whatman 3 MM filter paper for dried blood spots (DBS) for each participant, dried, and stored at room temperature until use.
DNA extraction
DBS were cut into small pieces with scissors and transferred into 2 mL microtubes. Genomic DNA was extracted using QIAamp DNA Mini blood kit according to the manufacturer’s protocol with some modifications. Briefly, 1 mL of PBS (1X)/saponin (0.5%), 180 µL of buffer ATL, 20 µL proteinase K, 200 µL of Buffer AL, 200 µL of ethanol (96–100%), 500 µL of Buffer AW1, 500 µL of Buffer AW2, and 200 µL of elution buffer (AE buffer) were added to each tube. The extraction was performed using the recommended incubation temperature and time with manufacturers’ centrifugation rate. Finally, the 1.5 mL Eppendorf tubes containing the extracted DNA were stored at − 20 °C until use.
Identification of Plasmodium species by real-time PCR
Plasmodium detection was performed by real-time PCR amplification of species-specific segments of the cytochrome b using the protocol described previously [28]. The assay was conducted to confirm and quantify P. vivax and mixed infection with P. falciparum since only these species were detected in the microscopic examination. The following nucleotide primers were used: forward-5′-TGCTACAGGTGCATCTCTTGTATTC-3′, and reverse-5′-ATTTGTCCCCAAGGTAAAACG-3′ specific for P. vivax, and forward-5′-ATGGATATCTGGATTGATTTTATTTATGA-3′, reverse-5′-TCCTCCACATATCCAAATTACTGC-3′ specific for P. falciparum. The PCR amplifications using CFX96™ Real-Time PCR detection system (Bio-Rad Laboratories Ltd) were carried out following 10 μL of molecular water, 4μL of Evagreen HRM Mix (5x) (Sotis biodyne), 0.5 μL of each primer, and 5 μL of DNA template with a final volume of 20 μL. The thermal cycling programs were 94 °C for 15 min (minutes) followed by 20 cycles of 30 s (seconds) at 94 °C, 58 °C for 1 min, 72 °C for one and a half min and 72 °C for 10 min followed by a cooling step of 20 °C. Negative (water) and positive (P. vivax DNA from blood sample with parasite density estimated ~ 8000 parasites/µL and P. falciparum DNA at 5 ng/µL) control samples were used in each PCR run. For each sample positive for P. vivax, the qPCR Ct value was estimated based on a threshold defined at 200.
Estimating parasite density from qPCR Ct value
The parasite densities per µL were estimated using qPCR for all 138 samples. However, to observe the correlation between qPCR ct values with microscopy results, the parasite densities per µL of fourteen randomly selected samples were determined on Giemsa-stained blood films. Blood films were read for 30 min under oil-immersion microscope by three experienced technicians (approximately 20,000 red blood cells). The correlation between qPCR Ct values (triplicate) with microscopy results (triplicate) was estimated using scatter diagrams and correlation coefficients (automatic weighted regression).
Duffy genotyping
Duffy genotypes were determined by PCR amplification from P. vivax human isolates since the DBS samples also contained human DNA. The primers for genotyping were forward-5′-GAGGCTTGTGCAGGCAGT-3′ and reverse-5′-CAAACAGCAGGGGAAATGAG-3′ specific for GATA-1 transcription factor binding site of the FY gene (− 33 T → C), and forward-5′-CCCTCAATTCCCAGGAGACT-3′ and reverse-5′-GCTGAGCCATACCAGAC ACA-3′ specific for exon 2 of the DARC gene (125 G → A) (Sigma Aldrich, France). The PCR was conducted in a 50 μL total PCR volume using 33 μL molecular water, 1 μL of each primer, 10 μL of 5X HOT blend Taq polymerase (Sotis biodyne), and 5 μL of DNA template, as previously described with minor modifications [29]. The PCR conditions were first denaturation at 94 °C for 15 min followed by 40 cycles at 94 °C for 20 s, 60 °C for 20 s, 72 °C for 1 min, and final elongation at 72 °C for 10 min. The PCR products were purified using Agencourt AMPure XP bead-based purification prior to sequencing. The purified PCR products were sent to Biofidal (France) for Sanger sequencing. The sequenced nucleotides were analyzed, using CLC Genomics Workbench 22.0 (Qiagen, Germany) software, on both strands. Sequences produced from the present study were aligned with their respective reference sequences.
Statistical analysis
All statistical analyses were performed with MedCalc software (version 20.014) and Statistical Package for Social Sciences (SPSS) version 25. Descriptive statistics were used to describe the characteristics of study participants, frequencies of Duffy genotypes, phenotypes, and other related variables using figures and tables. Outlier detection was used to detect anomalous observations in sample data by the Tukey test [30]. The outliers were categorized as ‘outside’ or ‘far out’ values. An outside value was defined as a value that is smaller than the lower quartile minus 1.5 times the interquartile range, or larger than the upper quartile plus 1.5 times the interquartile range. A far-out value was defined as a value that is smaller than the lower quartile minus 3 times the interquartile range, or larger than the upper quartile plus 3 times the interquartile range.
The regression analysis was used to define the relationship between asexual P. vivax parasitaemia estimated by microscopy and qPCR Ct value (defined by the best-fitting regression model), and then to deduce the parasitaemia from qPCR Ct values. The relationship was then confirmed with the paired t-test, intraclass correlation coefficient (ICC), and Passing-Bablok regression (bootstrap method). The independent non-parametric Mann–Whitney test was used to determine the differences between the median P. vivax parasitaemia related to Duffy antigen polymorphism with 95% confidence interval (CI).
The violin plot [31] was used to present the distribution of asexual P. vivax parasitaemia by Duffy antigen polymorphism. A P-value < 0.05 was considered statistically significant.
Results
Characteristics of the study population
Out of the total 151 recruited patients, 11 were P. falciparum positive(confirmed by two microscopists), and 2 were negative (confirmed by PCR). Thus, 138 symptomatic P. vivax infected patients were enrolled in this study; of which 135 were P. vivax single infections and the remaining three were mixed infections (P. vivax and P. falciparum) as determined by microscopy and real-time PCR. Thirty seven percent of the study participants were in the age group of 15–24 years old. The highest number (37%) of study participants were from Adama City Administration. The majority (66.7%) of study participants were males and were recruited during the data collection year of 2020 (67.4%) (Table 1).
Plasmodium vivax parasitaemia deduced by qPCR
Plasmodium vivax DNA was detected by qPCR amplification in all tested samples (N = 138). The median (range) Ct value was 19.07 (12.92–36.62). The outlier detection revealed 12 (8.7%) outside values in the sample set (Additional file 1: Fig. S1). The correlation between qPCR Ct values with microscopy results from the 14 randomly slected blood samples showed by the best-fitting regression model was estimated with the exponential curve as regression equation (Additional file 1: Fig. S2, Table S1). The ICC value indicated excellent consistency between microscopy and qPCR Ct values (ICC = 0.927; 95% CI 0.865–0.961). The paired t-test indicated that the estimated (from microscopy) and deduced (from qPCR Ct values) parasitaemia were not significantly different (mean difference 188.5 parasites/µL, 95% CI − 593.2 to 970.2, P = 0.629). The Passing–Bablok regression did not present evidence of a systematic or proportional bias. The estimations for the Intercept (95% CI) were − 9.92 (− 163.02 to 37.33), for the Slope (95% CI) 0.93 (0.84–1.14) and for the Spearman rank correlation coefficient (95% CI) 0.87 (0.78–0.93, P < 0.00001) (Additional file 1: Fig. S3).
The deduced median (range) P. vivax asexual parasitaemia of the 138 infected study participants was 4804(38–26,278) parasites/µL. We detected 12 (8.7%) samples with far-out values (≤ 110 parasites/µL) (Additional file 1: Fig. S4).
Duffy blood group genotyping
Duffy genotyping was successfully performed on 138 patients as shown in Table 2. The proportion of Duffy-negatives (FY*BES/FY*BES) in P. vivax infected patients was 2.9% (4/138). The duffy negatives were detected from Adama, Welenchity, Koka and Metehara (one from each site). Duffy genotype FY*B/FY*BES (48.6%) was the most common followed by FY*A/FY*BES genotype (25.4%). The FY*02 W.01/FY*02 N.01 genotype confering a weak expression of the Fyb antigen was observed in one patient. The FY*02 was the most frequently observed allele (65.9%) followed by FY*01 (42.8%). The most common (54.3%) phenotype detected in the study was Fy(a-b +) (Table 2).
Association between Duffy phenotypes and deduced P. vivax asexual parasitaemia
To determine whether and how Duffy phenotypes influence the asexual parasitaemia in P. vivax patients, this study analysed the relationship between the deduced parasite densities and the Duffy antigens distribution (deduced from Duffy genotypes). The median (range) P. vivax asexual parasitaemia of the 138 infected study participants by Duffy phenotypes are presented in the Table 3.
Plasmodium vivax infected Duffy-negative patients (FY*BES/FY*BES) showed a regularly low asexual parasitaemia (median, 53 parasites/µL) significantly lower (100-fold) than homogzygous (FY*A/FY*B, FY*A/FY*A or FY*B/FY*B) and heterozygous (FY*A/FY*BES, FY*B/FY*BES or FY*BES/FY*X) individuals (P < 0.0001). No significant difference in asexual P. vivax parasitaemia was observed between homozygous and heterozygous Duffy-positive individuals. Interestingly, four of the Duffy-negative patients were classified in the ≤ 110 parasites/µL group, while the proportions of patients carrying other Duffy phenotypes who had ≤ 110 parasites/µL were lower (5.7% [for FY*A/FY*BES]) and (3.0% [for FY*B/FY*BES]) (Fig. 2).
Discussion
Since the first discovery of Duffy blood groups in the 1950s [32], there have been many investigations focusing on its global distribution [33], its role in blood transfusion [34], and for P. vivax invasion pathway into red blood cells [35]. Later, the innate resistance of humans to P. vivax infections has been attributed to Duffy blood group polymorphisms [26]; however it is unclear whether Duffy blood group polymorphism confers either only partial or complete resistance against P. vivax. This is because of the detection of P. vivax infections [36] among the previously refractory Duffy antigen-negative red blood cells [11] as frequently observed in Africa over the last decade [13, 14, 19].
In the current study, P. vivax infection in Duffy-negative individuals was identified. However, the proportion (2.9%) of P. vivax-infected patients with Duffy-negatives was lower than previous studies conducted at different parts of Ethiopia (10.5–11.9%) [13]. Here, a larger number of FY*B/FY*BES (Fyb/Fyes) and FY*A/FY*BES (Fya/Fyes) carriers were observed among P. vivax infected patients in agreement with the previous study [19], indicating frequent heterozygous Duffy negativity of the study participants. This was in contrast with the predominant record of homogenous FY*BES in most African ethnic groups [10, 21], but confirmed the variation in the frequency of the Duffy genotypes across different populations. For instance, FY*B/FY*B was frequently observed in Eastern, while FY*B/FY*BES was in Southwestern Ethiopia [19]. A higher frequency of FY*B allele carriers was observed compared with the FY*A carriers. A consistent finding is that the probability of having the FY*B allele has increased in Africa [9, 19], in contrast to Asian countries where FY*A was the most frequent variant [25].
Phenotypically, the current results showed that the most common Duffy phenotype was Fy(a − b +) followed by Fy(a + b −) with a frequency of 54.3% and 30.5%, respectively, consistent with previous observations in neighbouring countries, such as Sudan [9]. Although all patients in the current study were clinical, the finding may suggest that individuals with Fy(a + b −) have a relatively low susceptibility to clinical P. vivax malaria compared to Fy(a − b +) due to diminishing effect of Fy(a + b −) on the binding of Duffy binding protein (PvDBP) at the erythrocyte surface, as it has been shown in Brazil [21]. The relatively more frequent observation of Fy(a − b +) phenotypes among P. vivax infected patients compared to healthy individuals in Sudan [9] also provided potential evidence for an association between Fy(a − b +) and its susceptibility to clinical malaria.
The FY*X (FY*02 W.01) allele encoded the weakly expressed Fyb antigen due to the reduced amount of Duffy protein caused by the mutation at position 89 (Arg > Cys) [37, 38]. Surprisingly, we observed for the first time that FY*X/FY*BES(FY*02 W.01/FY*02 N.01) genotype, which is not common in Africa [10], was detected from one P. vivax infected patient. The asexual P. vivax parasitaemia (5943 parasites/µL) detected in this patient was, however, similar to those found in homozygous and heterozygous Duffy-positives.
To date, it is well known that the asexual parasitaemia in patients with P. vivax malaria is influenced by several factors such as immunity, age, delay in seeking treatment, self-treatment behaviour before presentation, and other variety of host and parasite factors eventhough no significant differences were observed in the current study. Here, this study aimed at exploring an additional factor which is the effect of Duffy antigen polymorphisms that can also modulate asexual parasitaemia in P.vivax patients. The current study observed that heterozygous Duffy-positive individuals had similar range of parasite density with homozygous, as supported by previous findings in Ethiopia [13] and Thailand [39], but contrary to reports from Papua New Guinea suggesting that P. vivax parasitaemia was significantly lower in heterozygous than homozygous [22]. Particularly, Michon et al. showed the advantages of being heterozygous to reduce Duffy binding protein adherence to erythrocytes connecting with resistance against clinical P. vivax malaria [40].
However, the current study confirmed that the parasite density of P. vivax was significantly lower in Duffy-negative individuals than the Duffy-positives in line with previous studies elsewhere [9, 13, 22]. From this study, all Duffy-negative patients had parasitaemia ≤ 110 parasites/µL (median 53 parasites/µL), while Duffy-positive patients (homozygous or heterozygous) had an average parasitaemia between 1000 and 5000 parasites/µL. These data suggest two important features regarding vivax malaria epidemiology in Africa.
First, the very low parasite density observed in Duffy-negative patients might provide better estimates of the silent P. vivax parasites reservoir that may remain undetected by widely used microscopic examination although all P. vivax in Duffy negatives in the current study were detected by microscopy. It is important to note that the current frequent observation of vivax malaria among Duffy negative Africans might be due to the use of a more sensitive molecular tools. This suggests that P. vivax malaria was likely missed in Duffy-negative patients in endemic settings such as Africa where malaria diagnosis is routinely made by microscopy. This was supported by the previous findings which indicated that only four of the twenty P. vivax infections were microscopic-positive in Duffy-negative patients in other parts of Ethiopia [13]. This, therefore, further pointed out that P. vivax might be maintained in the Duffy-negative populations without clinical symptoms [41], and with the difficulty of microscopic examination [13], but could be transmitted between hosts [42].
This hypothesis is further substantiated by the fact that several studies noticed the high probability of transmission of P. vivax between hosts from submicroscopic individuals [43,44,45]. However, further study is needed to determine the relative contribution of Duffy antigen polymorphism in the transmissibility of P.vivax gametocytes to mosquitoes. The observation of P.vivax among Duffy-negative individuals, thus, might further complicate the poor understanding of the exact frequency of vivax malaria in Ethiopia and thereby across Africa where both Duffy-negatives and -positives co-exist.
Second, the very low parasite density found in Duffy-negative patients strongly suggests that the invasion pathway(s) used by P. vivax merozoites to invade Duffy-negative reticulocytes is (are) far less efficient compared to the classical pathway involving the interaction between the PvDBP and DARC in Duffy positive individuals. One hypothesis that is warranted to be envisioned is to consider that reticulocytes (or less mature erythroid cells from the bone marrow) of Duffy-negative subjects, which were thought to be devoid of the Duffy protein, express transitorily and weakly this protein at the course of erythroid cell differentiation. This hypothesis had already been explored by Dechavanne et al. [46], but needs to be confirmed and validated. However, this cannot reject the alternative hypothesis that P. vivax pathway used to invade Duffy negative reticulocytes is based on not very effective alternative interactions between unknown parasite ligand and reticulocytes receptors.
The limitation of this study includes failure to well describe the association of factors other than Duffy antigen polymorphism which could affect parasitaemia of P.vivax because of small samples. In addition, the number of Duffy negatives was too small for making firm conclusion regarding predictors of the parasite density.
In conclusion, the current study described the emerging perspectives on the relationship of Duffy antigen polymorphisms with parasite density of P. vivax. The complex relationships between hosts and parasites have clinical implications including the risk of being a silent reservoir, and thus needs introducing specific and sensitive diagnostic methods in areas where P. vivax malaria is endemic. Further investigations using longitudinal study and large samples (both clinical and community) from different parts of Ethiopia are needed to know the exact distribution of Duffy antigen and how Duffy antigen polymorphisms impact and shape the P. vivax malaria epidemiology in Ethiopia.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author on reasonable request. All relevant data are within the manuscript.
Abbreviations
- DARC:
-
Dufy antigen/receptor for chemokines
- RBCs:
-
Red blood cells
- SNP:
-
Single nucleotide polymorphism
- PCR:
-
Polymerase chain reaction
- PvDBP:
-
Plasmodium vivax Dufy binding protein
- PvRBP2b:
-
Plasmodium vivax Reticulocyte Binding Protein 2b
- TfR1:
-
Transferrin receptor 1
- WHO:
-
World Health Organization
References
WHO. World malaria report 2021. Geneva: World Health Organization; 2021.
Oboh MA, Oyebola KM, Idowu ET, Badiane AS, Otubanjo OA, Ndiaye D. Rising report of Plasmodium vivax in sub-Saharan Africa: implications for malaria elimination agenda. Sci Afr. 2020;10: e00596.
Auburn S, Cheng Q, Marfurt J, Price RN. The changing epidemiology of Plasmodium vivax: insights from conventional and novel surveillance tools. PLoS Med. 2021;18: e1003560.
Tadesse F, Fogarty AW, Deressa W. Prevalence and associated risk factors of malaria among adults in East Shewa Zone of Oromia Regional State, Ethiopia: a cross-sectional study. BMC Public Health. 2018;18:25.
Golassa L, White MT. Population-level estimates of the proportion of Plasmodium vivax blood-stage infections attributable to relapses among febrile patients attending Adama Malaria Diagnostic Centre, East Shoa Zone, Oromia, Ethiopia. Malar J. 2017;16:301.
De Silva JR, Lau YL, Fong MY. Genotyping of the Duffy blood group among Plasmodium knowlesi-infected patients in Malaysia. PLoS ONE. 2014;9: e108951.
Ntumngia FB, Thomson-Luque R, Torres LDM, Gunalan K, Carvalho LH, Adams JH. A novel erythrocyte binding protein of Plasmodium vivax suggests an alternate invasion pathway into Duffy-positive reticulocytes. MBio. 2016;7:e01261-16.
Chan LJ, Dietrich MH, Nguitragool W, Tham WH. Plasmodium vivax reticulocyte binding proteins for invasion into reticulocytes. Cell Microbiol. 2020;22: e13110.
Albsheer M, Pestana K, Ahmed S, Elfaki M, Gamil E, Ahmed SM, et al. Distribution of Duffy phenotypes among Plasmodium vivax infections in Sudan. Genes. 2019;10:437.
Höher G, Fiegenbaum M, Almeida S. Molecular basis of the Duffy blood group system. Blood Transfus. 2018;16:93–100.
Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks: the Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4.
Russo G, Faggioni G, Paganotti GM, Dongho GBD, Pomponi A, De Santis R, et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar J. 2017;16:74.
Lo E, Russo G, Pestana K, Kepple D, Abagero BR, Dongho GBD, et al. Contrasting epidemiology and genetic variation of Plasmodium vivax infecting Duffy-negative individuals across Africa. Intl J Infect Dis. 2021;108:63–71.
Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax–molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis. 2011;5: e1192.
Brazeau NF, Whitesell AN, Doctor SM, Keeler C, Mwandagalirwa MK, Tshefu AK, et al. Plasmodium vivax infections in Duffy-negative individuals in the Democratic Republic of the Congo. Am J Trop Med Hyg. 2018;99:1128–33.
Niangaly A, Gunalan K, Ouattara A, Coulibaly D, Sa JM, Adams M, et al. Plasmodium vivax infections over 3 years in Duffy blood group negative Malians in Bandiagara. Mali Am J Trop Med Hyg. 2017;97:744–52.
Oboh MA, Singh US, Ndiaye D, Badiane AS, Ali NA, Bharti PK, et al. Presence of additional Plasmodium vivax malaria in Duffy negative individuals from Southwestern Nigeria. Malar J. 2020;19:229.
Lo E, Yewhalaw D, Zhong D, Zemene E, Degefa T, Tushune K, et al. Molecular epidemiology of Plasmodium vivax and Plasmodium falciparum malaria among Duffy-positive and Duffy-negative populations in Ethiopia. Malar J. 2015;14:84.
Woldearegai TG, Kremsner PG, Kun JF, Mordmüller B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg. 2013;107:328–31.
Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol. 2018;34:420–9.
King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, et al. Fya/Fyb antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci USA. 2011;108:20113–8.
Kasehagen LJ, Mueller I, Kiniboro B, Bockarie MJ, Reeder JC, Kazura JW, et al. Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS ONE. 2007;2: e336.
Albuquerque SRL, de Oliveira CF, Sanguino EC, Tezza L, Chacon F, Castilho L, et al. FY polymorphisms and vivax malaria in inhabitants of Amazonas State. Brazil Parasitol Res. 2010;106:1049–53.
Chittoria A, Mohanty S, Jaiswal YK, Das A. Natural selection mediated association of the Duffy (FY) gene polymorphisms with Plasmodium vivax malaria in India. PLoS ONE. 2012;7: e45219.
Kano FS, de Souza AM, de Menezes TL, Costa MA, Souza-Silva FA, Sanchez BAM, et al. Susceptibility to Plasmodium vivax malaria associated with DARC (Duffy antigen) polymorphisms is influenced by the time of exposure to malaria. Sci Rep. 2018;8:13851.
Cavasini CE, de Mattos LC, Couto ÁADA, Couto VSDA, Gollino Y, Moretti LJ, et al. Duffy blood group gene polymorphisms among malaria vivax patients in four areas of the Brazilian Amazon region. Malar J. 2007;6:167.
Ba EH, Baird JK, Barnwell J, Bell D, Carter J, Dhorda M, et al. Microscopy for the detection, identification and quantification of malaria parasites on stained thick and thin blood films in research settings: procedure: methods manual. Geneva: World Health Organization; 2015.
Canier L, Khim N, Kim S, Sluydts V, Heng S, Dourng D, et al. An innovative tool for moving malaria PCR detection of parasite reservoir into the field. Malar J. 2013;12:1–12.
Roesch C, Popovici J, Bin S, Run V, Kim S, Ramboarina S, et al. Genetic diversity in two Plasmodium vivax protein ligands for reticulocyte invasion. PLoS Negl Trop Dis. 2018;2: e0006555.
Tukey JW. Exploratory data analysis. Pearson Modern Class. 1st ed. MA: Addison-Wesley; 1977.
Hintze JL, Nelson RD. Violin plots: a box plot-density trace synergism. Am Stat. 1998;52:181–4.
Cutbush M, Mollison P. The Duffy blood group system. Heredity. 1950;4:383–9.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266.
Compton A, Haber JM. The Duffy blood group system in transfusion reactions: a review of the literature and report of four cases. Blood. 1960;15:186–91.
Gai PP, Van Loon W, Siegert K, Wedam J, Kulkarni SS, Rasalkar R, et al. Duffy antigen receptor for chemokines gene polymorphisms and malaria in Mangaluru. India Malar J. 2019;18:328.
Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–71.
Dean L. Blood groups and red cell antigens. Bethesda: NCBI; 2005.
Meny G. The Duffy blood group system: a review. Immunohematology. 2010;26:51–6.
Pimpat Y, Leelawat K, Lek-Uthai U. Red blood cell duffy antigen receptor for chemokines and susceptibility to Plasmodium vivax infection in Thais,. Southeast Asian J Trop Med Public Health. 2016;47:885–9.
Michon P, Woolley I, Wood EM, Kastens W, Zimmerman PA, Adams JH. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. Febs Lett. 2001;495:111–4.
Niang M, Sane R, Sow A, Sadio BD, Chy S, Legrand E, et al. Asymptomatic Plasmodium vivax infections among Duffy-negative population in Kedougou, Senegal. Trop Med Health. 2018;46:45.
Commons RJ, Simpson JA, Thriemer K, Hossain MS, Douglas NM, Humphreys GS, et al. Risk of Plasmodium vivax parasitaemia after Plasmodium falciparum infection: a systematic review and meta-analysis. Lancet Infect Dis. 2019;19:91–101.
van Eijk AM, Sutton PL, Ramanathapuram L, Sullivan SA, Kanagaraj D, Priya GSL, et al. The burden of submicroscopic and asymptomatic malaria in India revealed from epidemiology studies at three varied transmission sites in India. Sci Rep. 2019;9:17095.
Vallejo AF, García J, Amado-Garavito AB, Arévalo-Herrera M, Herrera S. Plasmodium vivax gametocyte infectivity in sub-microscopic infections. Malar J. 2016;15:48.
Ferreira MU, Corder RM, Johansen IC, Kattenberg JH, Moreno M, Rosas-Aguirre A, et al. Relative contribution of low-density and asymptomatic infections to Plasmodium vivax transmission in the Amazon: pooled analysis of individual participant data from population-based cross-sectional surveys. Lancet Reg Health Am. 2022;9: 100169.
Dechavanne C, Dechavanne S, Metral S, Roeper B, Krishnan S, Fong R, et al. Duffy antigen expression in erythroid bone marrow precursor cells of genotypically Duffy negative individuals. BioRxiv. 2018;96: 508481.
Acknowledgements
The authors are grateful to Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Bahir Dar University, Bill and Melinda Gates Foundation, and European and Developing Countries Clinical Trials Partnership (EDCTP) for financial support of the study. Our special thanks also go to Malaria Genetics and Resistance Unit, Department of Parasites and Insect Vectors of Pasteur Institute, Paris, France for financial support for all molecular laboratory works. We would like to thank Tewabech Lemma and Tsehay Orlando (WHO certified microscopists at Adama Malaria Diagnostic Centre) for their laboratory support. We are indebted to all the study participants, and data collectors for their cooperation during the fieldwork.
Funding
This study was supported by the Bill and Melinda Gates Foundation (ID: INV-015996), European and Developing Countries Clinical Trials Partnership (EDCTP), and Malaria Genetics and Resistance Unit, Department of Parasites and Insect Vectors of Pasteur Institute, Paris, France.
Author information
Authors and Affiliations
Contributions
AA, DM and LG conceived and designed the study. AA collected the sample. AA and IB conducted the laboratory procedures. DM, LD and LG conducted the supervision. AA, IB, SM, CDL, DM and LG verifed, analysed, and interpreted the data. AA and IB drafted the original version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical clearance was obtained from the Institutional Review Board of Addis Ababa University, Aklilu Lemma Institute of Pathobiology (Ref. no. ALIPB IRB/18/2012/20), and National Research Ethics Review Committee (Ref. no. MoSHE/02/152/778/21). Study participants, their parents, or guardians had gotten detailed explanations about their rights, purpose, procedure, and benefit of the study and side effects of the procedure and written informed consent was also obtained. In addition, assent was obtained from children. The study participants were treated according to the national treatment guideline. All participants were assured of confidentiality throughout the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Summarized statistics of the regression equations (weighted least-squares regression) tested as the best-fitting regression model between P. vivax parasitemia estimated by microscopy and qPCR Ct values. Figure S1. Distribution of the qPCR Ct values of the 138 P. vivax blood samples collected from P.vivax infected patients, 2019–2021. The median (range) Ct value was 19.07 (12.92–36.62). The Outlier detection using the Tukey test revealed 12 (8.7%) outside values in the sample set (red dots). Figure S2. Scatter diagram of the best-fitting regression model (exponential curve as regression equation) between qPCR Ct values and microscopy results from the 14 blood samples. Figure S3. Passing–Bablok regression: scatter diagram and regression line of P. vivax parasitemia defined by microscopy and parasitemia deduced from qPCR Ct values. The Intercept (95% CI) were – 9.92 (− 163.02 to 37.33); the Slope (95% CI) 0.93 (0.84–1.14) and the Spearman rank correlation coefficient (95% CI) 0.87 (0.78–0.93, P < 10–5). Figure S4. Distribution of the P. vivax parasitemia deduced from qPCR Ct values. The median (range) P. vivax asexual parasitemia of the 138 infected study participants was 4804 (38–26,278) parasites/µL. The Tukey test (Outlier detection) detected 12 (8.7%) samples with far-out values (below 110 parasites/µL).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Abate, A., Bouyssou, I., Mabilotte, S. et al. Vivax malaria in Duffy-negative patients shows invariably low asexual parasitaemia: implication towards malaria control in Ethiopia. Malar J 21, 230 (2022). https://doi.org/10.1186/s12936-022-04250-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-022-04250-2