Abstract
Background
Prior to this project, only a handful of online visualizations existed for exploring the published literature on molecular markers of antimalarial drug resistance, and none specifically for the markers associated with Plasmodium falciparum resistance to the partner drugs in artemisinin-based combination therapy (ACT). Molecular information is collected in studies with different designs, using a variety of molecular methodologies and data analysis strategies, making it difficult to compare across studies. The purpose of this project was to develop a free online tool, which visualizes the widely published data on molecular markers of antimalarial drug resistance, starting with the two genes pfcrt and pfmdr-1, associated with resistance to the three most common partner drugs; amodiaquine, lumefantrine and mefloquine.
Methods
A literature review was conducted, and a standardized method was used to extract data from publications, and critical decisions on visualization were made. A global geospatial database was developed of specific pfmdr1 and pfcrt single nucleotide polymorphisms and pfmdr1 copy number variation. An informatics framework was developed that allowed flexibility in development of the tool over time and efficient adaptation to different source data.
Results
The database discussed in this paper has pfmdr1 and pfcrt marker prevalence information, from 579 geographic sites in 76 different countries, including results from over 86,000 samples from 456 articles published January 2001–May 2017. The ACT Partner Drugs Molecular Surveyor was launched by the WorldWide Antimalarial Resistance Network (WWARN) in March 2015 and it has attracted over 3000 unique visitors since then. Presented here is a demonstration of how the Surveyor database can be explored to monitor local, temporal changes in the prevalence of molecular markers. Here publications up to May 2017 were included, however the online ACT partner drug Molecular Surveyor is continuously updated with new data and relevant markers.
Conclusions
The WWARN ACT Partner Drugs Molecular Surveyor summarizes data on resistance markers in the pfmdr1 and pfcrt genes. The database is fully accessible, providing users with a rich resource to explore and analyze, and thus utilize a centralized, standardized database for different purposes. This open-source software framework can be adapted to other data, as demonstrated by the subsequent launch of the Artemisinin Molecular Surveyor and the Vivax Surveyor.
Similar content being viewed by others
Background
Malaria remains a major cause of morbidity and mortality with a concerning trend of increase in cases reported in the 2017 World Malaria Report (216 million cases of malaria in 2016, up 5 million from 2015) [1]. Africa still bears most of the burden with 90% of all malaria cases and deaths occurring in this region [1].
The continuing devastating impact of this disease is partly due to the emergence and spread of resistance to anti-malarials. Mutations in Plasmodium falciparum genes have long been established as markers of anti-malarial resistance [2,3,4,5], and the level of clinical treatment failure has been associated with the presence of these resistance mutations [6, 7]. Identification of both copy number variation (CNV) and single nucleotide polymorphisms (SNPs) in the P. falciparum genes allows monitoring of the emergence and spread of declining drug susceptibility in parasite populations [8].
With respect to the partner drugs used in currently recommended artemisinin-based combination therapy (ACT), resistance to amodiaquine has been associated with SNPs in both the P. falciparum chloroquine-resistance transporter (pfcrt) MAL7P1.27 and P. falciparum multidrug resistance gene 1 (pfmdr1) MAL5P1.230 [6, 7, 9] (Table 1). Increased risk of treatment failure after lumefantrine treatment [7] and decreased susceptibility to lumefantrine in vitro are associated with several specific alleles of pfmdr1 [10,11,12]. Resistance to mefloquine has been associated with amplification of the pfmdr1 gene [13].
Molecular surveillance can be used to estimate the impact of parasite resistance on preventative measures such as intermittent preventive therapy in pregnancy (IPTp) and seasonal malaria chemoprevention (SMC), as well as to assess the appropriateness of current recommended drug treatment policies [14,15,16]. One example of drug policy change having an impact on the prevalence of particular markers of resistance was observed in Tanzania, within 4 years from the introduction of artemether–lumefantrine (AL) as the first-line treatment in 2006, the prevalence of the pfmdr1 N86 and 184F alleles increased dramatically from 10% to 46% and < 10% to 40%, respectively [17]. Such evolution has direct consequences for patient treatment since the prevalence of a particular genotype in the parasite population is one factor considered in predicting an overall probability of treatment success [8].
It is presently still uncommon that treatment policies have been changed based on molecular studies alone, but two examples, from Mali and Tanzania, have demonstrated the use of local marker prevalences in evaluating drug policies [8, 18]. On a global level, mutations in P. falciparum dihydrofolate reductase (pfdhfr), PF3D7_0417200, and P. falciparum dihydropteroate synthase (pfdhps), PF3D7_0810800, markers of sulfadoxine-pyrimethamine (SP) resistance, played a role in the World Health Organization (WHO) decision to change IPTp policy recommendations from a 2-dose regimen of SP to SP administration at each scheduled antenatal care visit [19, 20].
The Millennium Development Goals Report 2015 highlights the importance of data collection and its use in evidence-based policymaking, recommending tailored local strategies and “opening up data… to provide free visualization and analysis tools” [21]. However, very few interactive maps that provide a summary of the anti-malarial drug resistance picture exist online, and none that allow the user to explore temporal trends. The London School of Hygiene and Tropical Medicine Africa map displays prevalence of mutations in the pfdhfr and pfdhps genes associated with SP resistance (http://www.drugresistancemaps.org). Another example is the Center for Global Development drug resistance maps which display the number of cases of CQ or SP resistance at the country level (http://www.cgdev.org/page/looking-drug-resistance). The more recently launched WHO Malaria Threats Map [22] presents information on resistance markers in the Pfkelch 13, pfcrt, pfmdr-1, and pfplasmepsin 2-3 genes (http://www.who.int/malaria/maps/threats). There are other examples of informative maps on malaria but these do not present drug resistance data (CDC interactive map (http://www.cdc.gov/malaria/map/index.html), and the Malaria Atlas Project (http://www.map.ox.ac.uk).
There is evidence of heterogeneity in marker prevalences between populations sampled from neighbouring regions within a country [23] which challenges the degree of spatial generalizability one can draw from a particular sample site. This highlights the need to make the most of all the data that do exist and supports the development of a single comprehensive source of all the evidence collected to summarize the data and to highlight gaps. This allows targeting of control programme resources to the areas of highest concern.
The WorldWide Antimalarial Resistance Network (WWARN) recognizes that data often need transforming into formats that are useful for a wide audience [24]. WWARN created an interactive map for point mutations in the pfdhfr and pfdhps genes (http://www.wwarn.org/dhfr-dhps-surveyor/). The next step was to summarize the published evidence on mutations in the pfcrt and pfmdr1 genes which have been associated with resistance to chloroquine (CQ) and the main ACT partner drugs, namely amodiaquine (AQ), lumefantrine (LUM), and mefloquine (MQ).
Collating the current evidence in a standardized approach is a key first step to facilitate the consideration of molecular marker prevalence evidence in informed treatment policies. The aim of this current effort was to conduct a periodic review of the published literature and create a useful, interactive visualization tool to summarize the information both spatially and temporally. WWARN’s intention is to present data without extrapolation or interpretation, using a clear, transparent methodology.
Methods
Data search and inclusion and exclusion criteria for publications
Studies have been identified every 6 months since 2015 through a PubMed literature review using the search terms ‘malaria AND (pfcrt OR pfmdr1 OR “molecular marker” OR “molecular markers”)’. The ACT partner drug Surveyor is regularly updated with newly published or unpublished data (minimum every 6 months). Articles published from 2001 onwards are included. Abstracts and text are scanned to determine which publications are relevant using the following inclusion criteria; reporting of at least one pfcrt or pfmdr1 genotype or haplotype from field isolates, i.e. not cultured strains (see Table 2 for a complete list of markers) and clear information on sample size, and location information (at least on country level). If malaria patient samples are assessed, only results from pre-treatment samples are considered. Details of the inclusion and exclusion criteria can be found in the Additional file 1. In this publication, data from articles published between January 2001 and May 2017 are presented. A full list of the publications captured in the search can be found in the Additional file 2. Future literature searches will also include prevalence data on plasmepsin2 CNV, related to piperaquine resistance [25, 26]. The ACT Partner Drug Molecular Surveyor is continuously updated with data from publications.
Data extraction and entry in the database
Data extracted from the publications includes marker genotype and haplotype prevalence (number of samples tested, number of samples with the pure and/or mixed genotype identified) and gene copy number as well as PubMedID and publication information, study year and geographic position (country, site, latitude and longitude). Data on multiple loci in pfmdr1 and pfcrt are extracted to determine the prevalence of genotypes and haplotypes (Table 2). If latitude/longitude were not reported in the article, geocoding was performed using online tools (http://www.gpsvisualizer.com/geocoder/ or http://www.google.com/maps).
An online web application has been developed to increase consistency in data entry and to support entry concurrently by multiple staff based anywhere in the world. Each user connects to the application through secure controlled access and each interaction with the application is audited. The main WWARN SQL database is a fault-tolerant database, which replicates data in different data centres to achieve high availability and durability. Daily back-ups are also applied in order to minimize data loss.
The following rules guide the process of data extraction and entry:
-
Data are extracted and entered per site per year, if this information is available.
-
Sample size must be specified per site and marker.
-
Articles are scanned particularly to identify reports of polyclonal infections. If polyclonal infections are present in the data set, they can be entered separately, or included in either the mutant and wild type categories or both, depending on the information given in the publication.
-
If no sample collection date is reported, the year of sample collection is estimated to 3 years prior to the publication year.
-
If the prevalence of only one allele (e.g. pfcrt 76T) is presented in the publication, the prevalence of the other allele (pfcrt K76) is calculated from the presented prevalence and extracted, if the mixed infections (pfcrt 76 K/T) were clearly accounted for.
-
If the marker prevalence is only presented in a graph in the article, the prevalence for each genotype is estimated based on the graph.
-
If only the prevalence of haplotypes (e.g. pfcrt 72–76) is presented in the publication, the prevalence of the single locus (pfcrt 76) is calculated from the haplotype prevalence, if all identified haplotypes were accounted for in the publication.
The online visualization tool
The Surveyor tool was built using the Google Web Toolkit, Bootstrap, a responsive front-end library and the WWARN maps surveyor open source framework (https://github.com/WorldwideAntimalarialResistanceNetwork/WWARN-Maps-Surveyor). This framework is a mapping platform for visualizing data to support research synthesis and reporting in global health settings and health research. The tool displays the prevalence of a molecular marker including mixed infections for both wild-type and mutant alleles, by location. Data can be filtered to view results on a particular ACT-partner drug, molecular marker, country, sample size, and sample collection year. Pin icons display the location of samples collected as reported in the publications and are coloured according to the prevalence of the marker being viewed. Pop-up boxes provide specific details about samples from a particular site. These boxes include a direct citation link to PubMed. If samples from more than 1 year have been reported from that site, a drop down menu allows each year to be viewed separately. The prevalence of molecular markers in two geographic sites or the same geographic site from different years can be compared with a tool below the map by selecting country, site and year from the drop-down menus. The full dataset can be downloaded as a single comma-separated value (csv) file which can be input for analysis in excel or other statistical software. The dataset that the user downloads can be customized to reflect the filter choices applied by the user.
Rules for data visualization:
-
For a study with multiple geographic sites, where the data cannot be separated by site, the data are displayed as all geographic sites together with the pin pointing at one of the sites.
-
If only the country of origin, but not the study site is specified, the data are displayed with a pin pointing at the capital of the country.
-
If a study collected samples over several years, the study is displayed with one pin coloured according to the data from the most recent year.
-
Inclusion or exclusion of mixed infections in the prevalence of a single genotype or haplotype is specified in the pop-up box.
Although there are 29 different loci (17 in pfcrt, and 12 in pfmdr1) captured in the database, the majority of the data concentrate on a few key loci; pfmdr1 codons 86, 184, 1246, and CNV; and pfcrt codons 72–76. To simplify visualization of the data on the Surveyor, only these key positions are listed in the marker list as they represent both the majority of the data, and the loci with the clearest evidence for a role in drug resistance. In addition to single nucleotide polymorphisms, haplotypes are also presented. The prevalence of particular single SNPs from data on haplotypes has been included in the single SNP prevalence. For example, if the user chooses ‘pfcrt 72–76 CXXXT’, where X represents any amino acid, then all publications that reported any haplotype will be displayed. In addition the prevalence of pfcrt 76T will be calculated based on all haplotypes presented in the article and displayed when the user chooses pfcrt 76T.
Analysis of the surveyor database
The Surveyor data set, openly available to download, can be used to provide visualization of spatial and temporal changes in marker prevalence over time. In the example presented in the paper, the data set was analysed to show the number of publications by year and region (Figs. 2, 3) as well two examples of temporal and spatial changes of marker prevalence. Firstly, temporal changes in the prevalence of pfcrt K76 and pfmdr1 N86 in a specific site, Tororo, Uganda, were addressed. The prevalence by year of the SNPs was calculated by dividing the sum of samples positive for a genotype (including mixed infections) for all studies in Tororo by the sum of tested samples (Fig. 5). Secondly, spatial and temporal changes of pfmdr1 N86 prevalence in six selected sub-Saharan countries were visualized (Fig. 6). The SNP prevalence from each of 42 studies was plotted by year and geographical site and colored by country. A line marked the mean prevalence of all study sites in a country by year. The mean was calculated by dividing the sum of samples positive for pfmdr1 N86 (including 86 N + Y) for all studies divided by the sum of tested samples by year, and presented based on the current first-line ACT in the country, according to WHO [2]. Selected sub-Saharan countries with data from > 50 samples in each site conducted between 2000 and 2015 were included. If studies were conducted over a range of years, the mean year was used. Countries were selected for analysis based on number of data points and spread over time. Data from Zanzibar were presented separately from Tanzania mainland since different first-line treatments were adopted in these areas. Fisher’s exact two-tailed test was used to compare the prevalence of the binned year groups 2003–2006 and 2009–2012. A full reference list of the original source publications used to create Fig. 6 can be found in Additional file 3. Graphs were created in Excel and Tableau version 10.5.
Results
The database
The database presented here included data from articles published between January 2001 and May 2017. The literature search identified 903 papers of which 450 (50%) were excluded (Fig. 1). Excluded studies fell into two groups; inappropriate study type and missing information. Within the study type category, most studies did not collect data of baseline prevalence in pfcrt and pfmdr1 markers (n = 249, 55%). This was expected as wide search terms were deliberately chosen to capture the majority of publications containing relevant data. Within the missing information category, 16 (4%) publications had unclear sample location details and six publications had an unspecified total number of samples analysed. Among the included studies 46 (10%) did not contain information on the year of sample collection. The sample collection year was then set to 3 years prior to publication. A total of 453 articles were included. In addition, one article was identified by personal communication and two articles in the PubMed literature search for the “SP Molecular Surveyor” since they included data on the ACT partner drug markers. Data have thus been extracted from 456 publications, which contained results from over 86,000 samples. A full list of the publications captured in the search can be found in the Additional file 2.
A range of 12 to 42 publications were included from each year between 2001 and May 2017 (Fig. 2). Samples had been collected between 1956 and 2017 and the median time between final sample collection and publication was 3 years, with a range of 0–50 years.
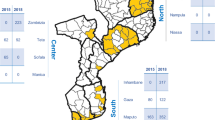
Studies were conducted in 76 different countries, covering a total of 579 geographic sites. Just over half of the studies reported data from at least one site in Africa (55% of publications) and 32% had at least one site in Asia (see Fig. 3). The vast majority of the publications (389, 86%) reported results for pfcrt with 141 reporting haplotypes for pfcrt72–76. A total of 330 papers (73%) reported any alleles in pfmdr1 and 87% of those (287) reported the pfmdr1 86 allele. Only 47 studies reported haplotypes for at least the three most commonly genotyped pfmdr1 loci (86/184/1246). Pfmdr1 copy number was reported in 84 (19%) publications.
The extracted data from all 456 publications are now presented as an online tool [27] entitled the WWARN ACT Partner Drugs Molecular Surveyor (Fig. 4). This interactive map allows users to explore the data visually using filters to select data by drug, marker, country, year of sampling, and sample size. Direct comparison between geographic sites, and at a single site between different years, can be undertaken in a special section below the main map.
Temporal and spatial trends of molecular markers
The Molecular Surveyor can be used to see changes over time in specific geographic sites or to visualize spatial and temporal trends of molecular marker prevalence on a local, national or global level. To illustrate the kinds of secondary studies that can be easily undertaken with the data freely available in the surveyor database, a detailed description of the data from Tororo, Uganda was prepared. Tororo was chosen, as it was the site in the database with the richest data set over time. Changes in the prevalence of markers pfmdr1 N86 and pfcrt K76 (both associated with resistance/tolerance to lumefantrine) were visualized, using samples spanning 2002 to 2015 (Fig. 5). These data demonstrate the power of bringing multiple datasets together as information is pooled from eight different publications including results from 3405 samples [28,29,30,31,32,33,34,35]. To investigate national trends, temporal pfmdr1 N86 prevalences were visualized in six sub-Saharan countries between 2000 and 2015 (Fig. 6). ACT was adopted as first-line therapy for uncomplicated malaria between 2003 and 2006 in sub-Saharan countries [36], which are presented by first-line treatment. The prevalence between binned years 2003–2006 and 2009–2012 were compared using Fisher’s exact test. The results suggest that pfmdr1 N86 prevalence increased significantly in all countries studied.
Prevalence of pfmdr1 N86 in sub-Saharan countries by study and year. Each point depicts one study, sized according to the number of tested samples. The line represents the annual mean of all studies in a country. Mixed infections 86 N + Y were included together with N86 in the analysis. The countries either have a AL as first-line treatment, b ASAQ as first-line treatment or c multiple first line treatments. This data was extracted from 42 source publications, details of which can be found in Additional file 3
Discussion
The aim is for the ACT Partner Drug Molecular Surveyor to be a dynamic tool updated with newly published data and new markers that will engage researchers. Additional information may need to be requested from investigators in order for the data set to be included in the Surveyor. The goal is that the surveyor can be a tool that will be useful to a broad range of users and data providers. This group could then work collaboratively to produce an increasingly integrated/coordinated surveyor system. Further development work may involve providing an even more detailed provision of information for local adjustment of optimal drug policies within a country.
Due to the large heterogeneity in reporting of molecular marker results, WWARN developed clear rules for extraction, including how to handle missing information, to complete the data extraction process. This standardized approach means that the resulting dataset is consistent and allow comparison between studies, although it also requires that some published data cannot be included due to the lack of reported details. Reporting of a minimum set of variables including sample size, study site, year of collection and how mixed infections are handled, would enable inclusion of more data in the future. Investigators are encouraged to include these data in each new manuscript, even if they have been specified in a previous publication.
Analysis of the Surveyor database can provide valuable insight in marker prevalence dynamics. As an example, and interestingly, molecular marker pfmdr1 N86 increased in six countries in sub-Saharan Africa between 2000 and 2016, independent of the adopted first-line treatment (Fig. 6). Overall, the data suggest there are factors other than/in addition to first-line treatment that govern prevalences and distributions of alleles. National, local and temporal policy variations of ACT deployment/use could occur. In the studied countries ACT use differed as much as from below 10% to above 80% [36]. The Surveyor data provides an opportunity for users to conduct their own analysis and link the data to ongoing research, as demonstrated by a 2018 paper from Okell et al. [37].
Conclusions
The Surveyor tool provides a unique resource for the research and policy communities, allowing the user to access summary information of thousands of samples from hundreds of geographic sites across the world, whilst still maintaining the depth of detail for those users who wish to investigate further a particular location. A key resource is the full data set that supports the tool and this can be freely downloaded. The dataset can be explored for local and national prevalence of resistance markers, and the rich resource of information it contains has been highlighted here. To date, the Surveyor has had over 3000 unique viewers.
Through developing this application, the WWARN software engineering team built a flexible open-source framework of over 200,000 lines of code that can be adapted for many different types of geospatial data. This learning was subsequently employed to launch two other surveyors. In April 2015, the Artemisinin Molecular Surveyor was launched, displaying data on the artemisinin resistance markers in the pfkelch 13 gene [38]. In 2016, the Vivax Surveyor was launched which summarizes the prevalence of chloroquine resistance in Plasmodium vivax [39].
One limitation of extracting data from the published literature is the inevitable delay between sample collection and publication date, making it hard to provide a really up-to-date combined picture of the evidence for resistance. Some of the benefits of developing this tool have been the clear identification of knowledge gaps, both geographically and temporally which could direct scarce resources to the regions of highest concern with respect to drug resistance surveillance and inform decisions on the use of drugs in preventive interventions.
Abbreviations
- ACT:
-
artemisinin based combination therapy
- Pfcrt :
-
P. falciparum chloroquine resistance transporter
- Pfmdr1 :
-
P. falciparum multidrug resistance transporter 1
- Pfpm :
-
P. falciparum plasmepsin
- Pfdhfr :
-
dihydrofolate reductase
- Pfdhps :
-
dihydropteroate synthase
- CNV:
-
copy number variation
- AL:
-
artemether–lumefantrine
- ASAQ:
-
artesunate–amodiaquine
- CQ:
-
chloroquine
- MQ:
-
mefloquine
References
WHO. World malaria report 2017. Geneva: World Health Organization. http://apps.who.int/iris/bitstream/10665/259492/1/9789241565523-eng.pdf?ua=1. Accessed Jan 2018.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–13.
Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8.
Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9.
Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–63.
Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, et al. Amodiaquine and artemether–lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–7.
Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P falciparum malaria after artemether–lumefantrine and artesunate–amodiaquine. Am J Trop Med Hyg. 2014;91:833–43.
Djimdé A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–1.
Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol. 2006;6:309–14.
Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether–lumefantrine in Africa. J Infect Dis. 2009;199:750–7.
Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–73.
Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob Agents Chemother. 2010;54:1200–6.
Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47.
Maiga H, Lasry E, Diarra M, Sagara I, Bamadio A, Traore A, et al. Seasonal malaria chemoprevention with sulphadoxine–pyrimethamine and amodiaquine selects pfdhfr-dhps quintuple mutant genotype in Mali. PLoS ONE. 2016;11:e0162718.
Venkatesan M, Alifrangis M, Roper C, Plowe CV. Monitoring antifolate resistance in intermittent preventive therapy for malaria. Trends Parasitol. 2013;29:497–504.
Grais RF, Laminou IM, Woi-Messe L, Makarimi R, Bouriema SH, Langendorf C, et al. Molecular markers of resistance to amodiaquine plus sulfadoxine–pyrimethamine in an area with seasonal malaria chemoprevention in south central Niger. Malar J. 2018;17:98.
Thomsen TT, Ishengoma DS, Mmbando BP, Lusingu JP, Vestergaard LS, Theander TG, et al. Prevalence of single nucleotide polymorphisms in the Plasmodium falciparum multidrug resistance gene (Pfmdr-1) in Korogwe District in Tanzania before and after introduction of artemisinin-based combination therapy. Am J Trop Med Hyg. 2011;85:979–83.
Mugittu K, Ndejembi M, Malisa A, Lemnge M, Premji Z, Mwita A, et al. Therapeutic efficacy of sulfadoxine-pyrimethamine and prevalence of resistance markers in Tanzania prior to revision of malaria treatment policy: Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase mutations in monitoring in vivo resistance. Am J Trop Med Hyg. 2004;71:696–702.
WHO Evidence Review Group. Intermittent preventive treatment of malaria in pregnancy (IPTp) with sulfadoxine–pyrimethamine (SP) Geneva: World Health Organization; 2012. http://www.who.int/malaria/mpac/sep2012/iptp_sp_erg_meeting_report_july2012.pdf. Accessed Jan 2018.
WHO. Updated WHO policy recommendation. Geneva: World Health Organization; 2012. http://www.who.int/malaria/iptp_sp_updated_policy_recommendation_en_102012.pdf. Accessed Jan 2018.
The Millennium Development Goals Report 2015. http://www.un.org/millenniumgoals/2015_MDG_Report/pdf/MDG/2015/rev(July1).pdf. Accessed Jan 2018.
WHO. Malaria Threats Map: tracking biological challenges to malaria control and elimination. Geneva: World Health Organization; 2018. http://apps.who.int/malaria/maps/threats/. Accessed Jan 2018.
Kavishe RA, Kaaya RD, Nag S, Krogsgaard C, Notland JG, Kavishe AA, et al. Molecular monitoring of Plasmodium falciparum super-resistance to sulfadoxine–pyrimethamine in Tanzania. Malar J. 2016;15:335.
Henry NL. Knowledge management: a new concern for public administration. Public Adm Rev. 1974;34:189.
Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis. 2017;17:174–83.
Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect Dis. 2017;17:164–73.
WWARN. ACT Partner Drug Molecular Surveyor 2015. http://www.wwarn.org/tracking-resistance/act-partner-drug-molecular-surveyor. Accessed May 2018.
Francis D, Nsobya SL, Talisuna A, Yeka A, Kamya MR, Machekano R, et al. Geographic differences in antimalarial drug efficacy in Uganda are explained by differences in endemicity and not by known molecular markers of drug resistance. J Infect Dis. 2006;193:978–86.
Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate–amodiaquine in Uganda. Antimicrob Agents Chemother. 2007;51:3023–5.
Tukwasibwe S, Mugenyi L, Mbogo GW, Nankoberanyi S, Maiteki-Sebuguzi C, Joloba ML, et al. Differential prevalence of transporter polymorphisms in symptomatic and asymptomatic falciparum malaria infections in Uganda. J Infect Dis. 2014;210:154–7.
Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, et al. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014;210:344–53.
Nankoberanyi S, Mbogo GW, LeClair NP, Conrad MD, Tumwebaze P, Tukwasibwe S, et al. Validation of the ligase detection reaction fluorescent microsphere assay for the detection of Plasmodium falciparum resistance mediating polymorphisms in Uganda. Malar J. 2014;13:95.
Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, et al. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg. 2014;91:54–61.
Nankabirwa JI, Conrad MD, Legac J, Tukwasibwe S, Tumwebaze P, Wandera B, et al. Intermittent preventive treatment with dihydroartemisinin-piperaquine in Ugandan schoolchildrens selects for Plasmodium falciparum transporter polymorphisms that modify drug sensitivity. Antimicrob Agents Chemother. 2016;60:5649–54.
Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, et al. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis. 2017;215:631–5.
WHO. Malaria country profiles. Geneva: World Health Organization; http://www.who.int/malaria/publications/country-profiles/en/. Accessed 23 Feb 2018.
Okell LC, Reiter LM, Ebbe LS, Baraka V, Bisanzio D, Watson OJ, et al. Emerging implications of policies on malaria treatment: genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether–lumefantrine and artesunate–amodiaquine in Africa. BMJ Glob Health. 2018;3:e000999.
WWARN. Artemisinin Molecular Surveyor 2015. http://www.wwarn.org/molecular-surveyor-k13. Accessed 21 Dec 2018.
Commons RJ, Thriemer K, Humphreys G, Suay I, Sibley CH, Guérin PJ, et al The Vivax Surveyor: online mapping database for Plasmodium vivax clinical trials. Int J Parasitol Drugs Drug Resist. 2017;7:181–90.
Authors’ contributions
SD and GH drafted the manuscript. IS, SG, and NT built and developed the application. SD and GH decided on the rules for data extraction and display. SD, GH and SS carried out the literature search and data extraction. AB edited the manuscript and figures. All authors read and approved the final manuscript.
Acknowledgements
Thanks to the investigators who conducted the studies presented on the Surveyor and the WorldWide Antimalarial Resistance Network (WWARN) team for technical and administrative support. Thanks to Carol Hopkins Sibley for valuable comments and suggestions on the draft manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset analyzed for the summary reported here are available to download from the WWARN website within the ACT Partner Drug Surveyor tool.
Funding
WWARN is funded by a Bill and Melinda Gates Foundation grant and the Exxon Mobil Foundation. The funders did not participate in developing the protocol or writing the paper.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1.
Inclusion and exclusion criteria for the literature review.
Additional file 2.
All studies in the Surveyor Jan 2001–May 2017.
Additional file 3.
All source publications for the data presented in Fig. 6.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Otienoburu, S.D., Suay, I., Garcia, S. et al. An online mapping database of molecular markers of drug resistance in Plasmodium falciparum: the ACT Partner Drug Molecular Surveyor. Malar J 18, 12 (2019). https://doi.org/10.1186/s12936-019-2645-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-019-2645-x