Abstract
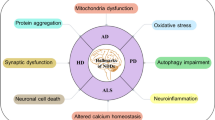
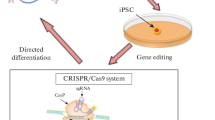
The review analyzes the results of modern studies that involve gene therapy methods for the correction of neurodegenerative diseases. Approaches based on gene-editing technologies and their prospective use in medicine are considered. There may be limitations to the use of the genome-editing tools in the treatment of these pathologies due to features of the pathogenesis of neurodegenerative diseases.


Similar content being viewed by others
REFERENCES
Bannikov, A.V. and Lavrov, A.V., CRISPR/CAS9, the king of genome editing tools, Mol. Biol. (Moscow), 2017, vol. 51, no. 4, pp. 514–525.
Baranov, V.S., Molecular medicine: molecular diagnostics, preventive medicine, and gene therapy, Mol. Biol. (Moscow), 2000, vol. 34, no. 4, pp. 590–600.
Vetchinova, A.S., Illarioshkin, S.N., Novosadova, E.V., et al., CRISPR/CAS9 artificial nuclease system as a tool for studying monogenic forms of Parkinson’s disease, Sib. Med. Obozr., 2017, no. 4, pp. 53–58.
Vetchinova, A.S., Konovalova, E.V., Volchkov, P.Yu., et al., Genome editing on a cellular model of the genetic form of Parkinson’s disease, Geny Kletki, 2016, vol. 9, no. 2, pp. 114–118.
Vetchinova, A.S., Konovalova, E.V., Lunev, E.A., and Illarioshkin, S.N., Genome technology editing and its possible use in cell neuroscience, Ann. Nevrol., 2015, vol. 9, no. 4, pp. 59–64.
Malakhova, A.A., Sorokin, M.A., Sorokina, A.E., et al., Using genome editing techniques to create isogenic cell lines modeling the Huntington’s disease in vitro, Geny Kletki, 2016, vol. 11, no. 2, pp. 106–113.
Smirnov, A.V., Yunusova, A.M., Lukyanchikova, V.A., and Battulin, N.R., CRISPR/Cas9, a universal tool for genomic engineering, Russ. J. Genet.: Appl. Res., 2017, vol. 7, no. 4, pp. 440–458.
An, M.C., O’Brien, R.N., Zhang, N., et al., Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system, PLoS Curr., 2014, vol. 6. https://doi.org/10.1371/currents.hd.0242d2e7ad7222-5efa72f6964589369a
Calatayud, C., Carola, G., Consiglio, A., and Raya, A., Modeling the genetic complexity of Parkinson’s disease by targeted genome edition in iPS cells, Curr. Opin. Genet. Dev., 2017, vol. 46, pp. 123–131.
Cermak, T., Doyle, E.L., Christian, M., et al., Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting, Nucleic Acids Res., 2011, vol. 39. e82.
Choi, W., Kim, E., Yum, S.Y., et al., Efficient PRNP deletion in bovine genome using gene-editing technologies in bovine cells, Prion, 2015, vol. 9, pp. 278–291.
Chugunova, A.A., Dontsova, O.A., and Sergiev, P.V., Methods of genome engineering: a new era of molecular biology, Biochemistry (Moscow), 2016, vol. 81, pp. 662–677.
Doudna, J.A. and Charpentier, E., Genome editing. The new frontier of genome engineering with CRISPR-Cas9, Science, 2014, vol. 346, p. 1258096.
Ertekin-Taner, N., Genetics of Alzheimer’s disease: a centennial review, Neurol. Clin., 2007, vol. 25, pp. 611–667.
Feng, W., Liu, H.-K., and Kawauchi, D., CRISPR-engeneered genome editing for the next generation of neurological disease modeling, Progr. Neuropsychopharmacol. Biol. Psychiatry, 2018, vol. 81, pp. 459–467.
Fink, K.D., Deng, P., Gutierrez, J., et al., Allele-specific reduction of the mutant Huntington allele using transcription activator-like effectors in human Huntington’s disease fibroblasts, Cell Transpl., 2016, vol. 25, pp. 677–686.
Fong, H., Wang, C., Knoferle, J., et al., Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells, Stem Cell Rep., 2013, vol. 1, pp. 226–234.
Gyorgy, B., Ingelsson, M., Loov, C., et al., CRISPR-Cas9 mediated gene editing in a monogenic form of Alzheimer’s disease, Mol. Ther., 2016, vol. 24, pp. S226–S227.
György, B., Lööv, C., Zaborowski, M.P., et al., CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease, Mol. Ther. Nucl. Acids, 2018, vol. 11, pp. 429–440.
Hallmann, A.-L., Araúzo-Bravo, M.J., Mavrommatis, L., et al., Astrocyte pathology in a human neural stem cell model of frontotemporal dementia caused by mutant TAU protein, Sci. Rep., 2017, vol. 7, p. 42991.
Hsu, P.D., Lander, E.S., and Zhang, F., Development and applications of CRISPR-Cas9 for genome engineering, Cell, 2014, vol. 157, pp. 1262–1278.
Jeon, Y., Choi, Y.H., Jang, Y., et al., Direct observation of DNA target searching and cleavage by CRISPR-Cas12a, Nat. Commun., 2018, vol. 9, p. 2777.
Jo, Y.I., Kim, H., and Ramakrishna, S., Recent developments and clinical studies utilizing engineered zinc finger technology, Cell. Mol. Life Sci., 2015, vol. 72, pp. 3819–3830.
Kavli Prize in Nanoscience 2018. http://kavliprize.org/sites/default/files/%25nid%25/prize_overvi-ew_attachemnts/TKP%202018%20Nanoscience_citation.pdf.
Kim, Y.G., Cha, J., and Chandrasegaran, S., Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain, Proc. Natl. Acad. Sci. U.S.A., 1996, vol. 93, pp. 1156–1160.
King, A., A model challenge, Nature, 2018, vol. 559, pp. S13–S15.
Klein, C. and Westenberger, A., Genetics of Parkinson’s disease, Cold Spring Harbor Perspect. Med., 2012, vol. 2, p. a008888.
Kolli, N., Lu, M., Maiti, P., et al., Application of the gene editing tool, CRISPR-Cas9, for treating neurodegenerative diseases, Neurochem. Int., 2018, vol. 112, pp. 187–196.
Komor, A.C., Kim, Y.B., Packer, M.S., et al., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature, 2016, vol. 533, pp. 420–424.
Lao, Y.H., Li, M., Gao, M.A., et al., HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or Natronobacterium gregoryi argonaute, Adv. Sci., 2018, vol. 5, p. 1700540.
Leone, P., Shera, D., McPhee, S.W., et al., Long-term follow-up after gene therapy for canavan disease, Sci. Transl. Med., 2012, vol. 4, p. 165ra163.
Li, X., Wang, Y., Liu, Y., et al., Base editing with a Cpf1-cytidine deaminase fusion, Nat. Biotechnol., 2018, vol. 36, pp. 324–327.
Li, X., Yu, B., Sun, Q., et al., Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons, Proc. Natl. Acad. Sci. U.S.A., 2017, vol. 115, no. 2, pp. 415–420. http://www.pnas.org/cgi/doi/10.1073/pnas.1703601115
Lin, F.B., Liu, X., Xie, J.W., et al., Verification of a sporadic Alzheimer disease model in SORL1 gene knockout mice, Nan Fang Yi Ke Da Xue Xue Bao, 2018, vol. 38, pp. 289–295.
Lykken, E.A., Shyng, C., Edwards, R.J., et al., Recent progress and considerations for AAV gene therapies targeting the central nervous system, J. Neurodev. Disord., 2018, vol. 10, p. 16.
Ma, H., Marti-Gutierrez, N., Park, S.W., et al., Correction of a pathogenic gene mutation in human embryos, Nature, 2017, vol. 548, pp. 413–419.
Malankhanova, T.B., Anastasia, A., Malakhova, A.A., et al., Modern genome editing technologies in Huntington’s disease research, J. Huntington’s Dis., 2017, vol. 6, pp. 19–31.
Mendell, J.R., Al-Zaidy, S., Shell, R., et al., Single-dose gene-replacement therapy for spinal muscular atrophy, New Engl. J. Med., 2017, vol. 377, pp. 1713–1722.
Miller, J.C., Tan, S., Qiao, G., et al., A TALE nuclease architecture for efficient genome editing, Nat. Biotechnol., 2011, vol. 29, pp. 143–148.
Mittermeyer, G., Christine, C.W., Rosenbluth, K.H., et al., Long-term evaluation of a phase 1 study of AADC gene therapy for Parkinson’s disease, Hum. Gene Ther., 2012, vol. 23, pp. 377–381.
Nimsanor, N., Poulsen, U., Rasmussen, M.A., et al., Generation of an isogenic, gene-corrected iPSC line from a symptomatic 59-year-old female patient with frontotemporal dementia caused by a R406W mutation in the microtubule associated protein tau (MAPT) gene, Stem Cell Res., 2016, vol. 17, pp. 576–579.
Pihlstrøm, L., Wiethoff, S., and Houlden, H., Genetics of neurodegenerative diseases: an overview, Handb. Clin. Neurol., 2017, vol. 145, pp. 309–323. https://doi.org/10.1016/B978-0-12-802395-2.00022-5
Powell, S.K., Gregory, J., Akbarian, S., and Brennand, K.J., Application of CRISPR/Cas9 to the study of brain development and neuropsychiatric disease, Mol. Cell. Neurosci., 2017, vol. 82, pp. 157–186.
Ring, K.L., An, M.C., Zhang, N., et al., Genomic analysis reveals disruption of striatal neuronal development and therapeutic targets in human Huntington’s disease neural stem cells, Stem Cell Rep., 2015, vol. 5, pp. 1023–1038.
Sasaguri, H., Nagata, K., Sekiguchi, M., et al., Introduction of pathogenic mutations into the mouse Psen1 gene by Base Editor and Target-AID, Nat. Commun., 2018, vol. 9, p. 2892.
Savell, K.E. and Day, J.J., Application of CRISPR/Cas9 in the mammalian central nervous system, Yale J. Biol. Med., 2017, vol. 90, pp. 567–581.
Savitskaya, E.E., Musharova, O.S., and Severinov, K.V., Diversity of CRISPR-cas prokaryotic adaptive immunity mechanisms and applications for biotechnology, Biochemistry (Moscow), 2016, vol. 81, no. 7, pp. 870–880.
Schaub, R.T., Anders, D., Golz, G., et al., Serum nerve growth factor concentration and its role in the preclinical stage of dementia, Am. J. Psychiatry, 2002, vol. 159, pp. 1227–1229.
Schmid, B. and Haass, C., Genomic editing opens new avenues for zebrafish as a model for neurodegeneration, J. Neurochem., 2013, vol. 127, pp. 461–470.
Shimizu, Y., Sollu, C., and Meckler, J.F., Adding fingers to an engineered zinc finger nuclease can reduce activity, Biochemistry, 2011, vol. 50, pp. 5033–5041.
Shin, J.W., Kim, K. H., Chao, M.J., et al., Permanent inactivation of Huntington’s disease mutation by personalized allelespecific CRISPR/Cas9, Hum. Mol. Genet., 2016, vol. 25, pp. 4566–4576.
Soldner, F., Laganiere, J., Cheng, A.W., et al., Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations, Cell, 2011, vol. 146, pp. 318–331.
Soldner, F., Stelzer, Y., Shivalila, C.S., et al., Parkinson-associated risk variant in enhancer element produces subtle effect on target gene expression, Nature, 2016, vol. 533, pp. 95–99.
Stavrovskaya, A.V., Novosadova, E.V., Olshansky, A.S., et al., Effect of cell genome editing on the outcome of neurotransplantation in experimental Parkinsonism, Sovrem. Tehnol. Med., 2017, no. 9, pp. 7–14.
Stepanichev, M.Yu., Current approaches and future directions of gene therapy in Alzheimer’s disease, Neurochem. J., 2011, vol. 5, pp. 159–169.
Suzuki, K. and Belmonte, J.C., In vivo genome editing via the HITI method as a tool for gene therapy, J. Hum. Genet., 2018, vol. 63, pp. 157–164.
Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., et al., In vivo genome editing via CRISPR/Cas9 mediated homologyindependent targeted integration, Nature, 2016, vol. 540, pp. 144–149.
Tosolini, A.P. and Sleigh, J.N., Motor neuron gene therapy: lessons from spinal muscular atrophy for amyotrophic lateral sclerosis, Front. Mol. Neurosci., 2017, vol. 10, p. 405.
Tuszynski, M.H., Thal, L., Pay, M., et al., A phase 1 clinical trial of nerve growth factor gene therapy for Alzheimer disease, Nat. Med., 2005, vol. 11, pp. 551–555.
Tuszynski, M.H., Yang, J.H., Barba, D., et al., Nerve growth factor gene therapy: activation of neuronal responses in Alzheimer disease, J.A.M.A. Neurol., 2015, vol. 72, pp. 1139–1147.
Uemura, T., Mori, T., Kurihara, T., et al., Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in uteroelectroporation techniques, Sci. Rep., 2017, vol. 6, p. 35861.
Verheijen, J. and Sleegers, K., Understanding Alzheimer disease at the interface between genetics and transcriptomics, Trends Genet., 2018, vol. 34, pp. 434–447.
Wang, X., Wang, Y., Wu, X., et al., Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors, Nat. Biotechnol., 2015, vol. 33, pp. 175–178.
Woodruff, G., Young, J.E., Martinez, F.J., et al., The presenilin-1 ΔE9 mutation results in reduced γ-secretase activity, but not total loss of PS1 function, in isogenic human stem cells, Cell Rep., 2013, vol. 5, pp. 974–985.
Wright, A.V., Nunez, J.K., and Doudna, J.A., Review biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering, Cell, 2016, vol. 164, pp. 29–44.
Xu, S., Cao, S., Zou, B., et al., An alternative novel tool for DNA editing without target sequence limitation: the structure-guided nuclease, Genome Biol., 2016, vol. 17, p. 186.
Xu, T., Li, Y., van Nostrand, J.D., et al., Cas9-based tools for targeted genome editing and transcriptional control, Appl. Environ. Microbiol., 2014, vol. 80, pp. 1544–1552.
Yee, J.K., Off-target effects of engineered nucleases, FEBS J., 2016, vol. 283, pp. 3239–3248.
Zhou, X., Xin, J., Fan, N., et al., Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer, Cell. Mol. Life Sci., 2015, vol. 72, pp. 1175–1184.
Funding
This study was supported by the Program of the Presidium of RAS “Fundamental Science for Biomedical Technologies”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by M. Stepanichev
Rights and permissions
About this article
Cite this article
Stepanichev, M.Y. Prospects for the Use of Genome-Editing Technology to Correct Neurodegenerative Diseases. Adv Gerontol 9, 154–163 (2019). https://doi.org/10.1134/S2079057019020218
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079057019020218