Abstract
Ceratitis capitata (Wiedemann) is a polyphagous plant pest organism causing serious damage on more than 300 fruit types. Although the medfly is present in Europe, it is so far not established in Germany. In the years 2015–2017, an official survey was conducted by the German Federal Plant Protection Organisations to elucidate the current occurrence of this organism in Germany. While in 2015 only small numbers of flies were captured in traps situated in all German Federal States, a peak of 188 specimens could be trapped in 2016 with a decline to 29 specimens in 2017. Highest abundance of the fruit fly was detected in Southern Germany and at a single spot east of Berlin. The present study aimed to identify the origin of the trapped fruit flies and to assess the general potential of the species for overwintering in Germany. Using two approaches with cytochrome oxidase subunit I (COI), Sanger barcoding sequencing first and subsequent analysis of ten small simple repeat microsatellite loci from literature, the worldwide spread of C. capitata describing three phases could be retraced by analysing samples from Germany and 11 other countries. German and French populations were found to build the most recent stage of this species distribution. Specimens trapped in Germany obviously resulted from different introduction events. They most strongly resembled populations from France and Croatia but were not identical to these populations. We found strong evidence that specimens captured between 2016 and 2017 could not overwinter in Germany.
Similar content being viewed by others
Introduction
Ceratitis capitata (Wiedemann, Tephritidae; common name medfly) is a polyphagous and multivoltine pest species. C. capitata infests more than 300 commercially grown fruit types including citrus, stone fruits, pome fruits, tomatoes, figs and others (Meats and Smallridge 2007; Morales et al. 2004). The economic damage is caused by larval trophic activity in the fruit and the transmission of bacteria and fungi through the adult flies (Cayol and Causse 1993). The medfly has a short life cycle, e.g. only 8 days of larval development in immature peaches as one of the mainly infested fruits in the Mediterranean distribution area of the fruit fly (Fimiani 1989). The whole generation cycle is temperature sensitive and generally finished within 23 days at temperatures between 13 and 28 °C (CABI/EPPO 1997); live span of adults is usually up to 3 months. In consequence, the medfly produces up to 13 generations per year in laboratory lineages and more than 11 generations in its sub-Saharan origin (Fimiani 1989). In the Mediterranean area, the medfly develops at least six generations per year (Procedia/ Naples) with two to three generations in the northern parts of Italy (Bologna, Po Valley). In several cases under inappropriate temperature conditions, one generation cycle took over 250 days in maximum value and approached or even exceeded 200 days in average terms (Rigamonti 2004). C. capitata shows no diapause in the tropical habitats as long as suitable food sources are available, whereas it shows a diapause of several months in its Mediterranean habitats. In these areas, the medfly overwinters in larval stages on citrus fruits. In the northern part of Italy (Lombardy), it commonly can survive as adults in indoor environments with 10–12 °C (Rigamonti 2004). In central Europe, the fruit fly is regarded as a secondary pest with transient pest status with no overwintering observed so far, as temperatures get too low during the winter. C. capitata possesses a high intrinsic rate for its population increase following the ecological scheme of r-strategy that is characterised by fast infestation of new host fruit types and areas accompanied by a high fecundity (Fletcher 1989). Compared to other fruit flies, i.e. Dacus spp., dispersal over long distances is not an important adaptive feature of C. capitata (Fletcher 1989). Nevertheless migratory movements of over 20 km have been reported but most re-trapping experiments using laboratory reared labelled male flies showed movements about distances of only several hundred metres (Fletcher 1989).
In the past 200 years, C. capitata has spread from its supposed origin in tropical Africa (Kenya) to a number of tropical and subtropical countries all over the world (Malacrida et al. 1998). This dissemination was mainly driven by trade of fruits and other products infested mostly by larval stages. The earliest appearance of C. capitata in the Mediterranean basin is reported for the southeast of Spain (Malaga, 1842) and parts of Northern Africa (Morocco) followed by derived populations in Tunisia and Egypt (Malacrida et al. 1998). The main hosts in these countries are peaches (Spain), apples, pears (Tunisia), and guava as well as citrus fruits in Egypt (mandarin, navel orange). Spread to Italy and France occurred during the second half of the nineteenth century, and spread to countries at the Balkan peninsula (former Yugoslavia) was reported relatively late in 1947 (Fimiani 1989; Fig. 6a).
In temperate Europe, only transient populations of C. capitata have been observed, resulting from several introduction events. In France, the medfly was first reported 1900, disappeared for a decade and was re-introduced 1915 (Fimiani 1989). In Germany, infestations of the fruit fly were reported first in 1934 in Frankfurt (Main) and then C. capitata re-appeared several times in the subsequent decades. Past outbreaks in Germany were mainly described from Baden-Wuerttemberg, which is situated in the South of Germany and has mainly a milder climate than other regions (Fischer-Colbrie and Busch-Petersen 1989).
When introduced pests cannot be traced back directly to an imported consignment, differences in biochemical or genetic characteristics could be used to elucidate population dynamics. As early as in the 1970s and 1980s, genetic variation of C. capitata was investigated by means of enzyme isoelectric focusing and multilocus enzyme analysis (MLEE) covering differences in structure and composition of enzyme proteins of at least 25 different enzymes/biochemical loci (Milani et al. 1989). In 2000, Bonizzoni et al. (2000) designed an approach to investigate genetic variation between medfly populations using 10 neutral microsatellite markers (SSR-markers) providing a higher genetic resolution compared to MLEE. The study presented here aims to determine the origin of C. capitata trapped during an official survey conducted in the federal states of Germany during the years 2016 and 2017. The population structure of the sampled individuals was analysed using the approach of Bonizzoni et al. (2000). German medflies were compared with individuals trapped in other states in Europe (France, Spain, Italy and Croatia), in Northern Africa (Tunisia, Egypt), Oriental Mediterranean area (Lebanon), South America (Brazil) and Africa (Cameroon, Togo, Republic of South Africa) to elucidate pathways in migration. Additionally, medflies from two years of sampling (2016 and 2017) trapped in Germany were compared for genetic variation to determine, whether at least short-term establishment might occur in some parts of Germany, e.g. in mild winters.
Materials and methods
Official survey and acquisition of reference material
An official survey, carried out by the German Plant Protection Services under supervision of the Julius Kühn Institute (JKI)–Federal Research Centre for Cultivated Plants, was initiated in the timespan between 2015 and 2017 to elucidate the current infestation status of C. capitata in Germany. For this purpose, a number of fruit orchards mainly growing apple trees (Malus domesticus) but also mixed cultures of apple, cherry and plum trees were monitored. For baiting C. capitata specifically, attractant traps using a pheromone dispenser in combination with an insecticide (Easy Trap®, Sorygar, Spain) were prepared containing the male attractant-related baiting agent (male lure) trimedlure. In the years 2015 and 2016, 214 and 247 attractant traps were installed, respectively, while in 2017, 133 traps were used for baiting the medfly (Fig. 1). Within orchards, a trap density of one trap per 2 km2 was envisaged to achieve optimal survey intensity in regard to main disposal of males only several 100 m away from the place of pupation. Traps were installed in 14, 13 and 7 of the German federal states for the different years. A small number of sites was monitored continuously over the three years. Trapped specimens were stored in 95% ethanol after weekly or biweekly collection of the traps. Morphological determination of the fruit flies was done by the inspectors of the Federal Plant Protection Services or by JKI at least for confirmation of the identity.
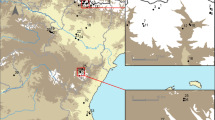
Results of the official survey of medflies in Germany. Trap count and trapped specimens are indicated by colour for different Federal States (a), and course of trapped individuals over the months shown by blue line diagrams (b) for each year of the official survey. Federal States are abbreviated by two character codes: BW—Baden-Wuerttemberg; BY—Bavaria; BE—Berlin; BB—Brandenburg; HH—Hamburg; HB—Bremen; HE—Hesse; MV—Mecklenburg-Western Pomerania; NI—Lower Saxony; NW—North Rhine-Westphalia; RP—Rhineland-Palatinate; SL—Saarland; SN—Saxony; ST—Saxony-Anhalt; SH—Schleswig–Holstein; TH—Thuringia
Following the distribution pathways of C. capitata, a number of individuals from reference populations of the medfly were acquired from researchers and Plant Protection Services (see acknowledgement). Reference material was chosen to represent and follow the pathways of spread of the medfly as stated in the literature (e.g. Barr 2009; Bonizzoni et al. 2000; Malacrida et al. 1998). Reference material therefore comprised samples from areas close to the origin in sub-Saharan Africa to the countries in the North African and European Mediterranean habitat to South America and recent outbreaks in France (Table 2).
All of the laboratory work for DNA extraction and PCR amplification of the cytochrome oxidase subunit I (COI) barcoding region as well as analysis of loci for population genetics including amplified fragment scoring was conducted in sub-contract by MWG-Eurofins Applied Genetics department/Eurofins Medigenomix GmbH (Ebersberg, Germany; see acknowledgements) under authority of the Julius Kühn Institute.
Meteorological data
Meteorological data comprising daily maximum, minimum and mean temperature of the relevant years 2015–2017 were taken from the database of the German forecast service Deutscher Wetterdienst (Offenbach, Germany). A subset of six meteorological stations was chosen to represent the areas in Germany where most of the medflies were trapped. Therefore, temperature data of the stations situated in Mainz (Rhineland-Palatinate), Konstanz (Baden-Wurttemberg), Würzburg (Bavaria), Manchnow (Brandenburg) close to Frankfurt (Oder) and Dresden-Klotzsche (Saxony) were depicted. Spearman rank correlation analysis was calculated for the number of days showing a maximum temperature below 0 °C, the years 2015–2017, and the places of meteorological stations. Spearman rank correlation diagrams were performed using the varclus function for variable clustering as implemented in R-package Hmisc (Harrell 2021). Wilcoxon’s signed rank tests for the number of days with maximum temperature below 0 °C between the different years and stations, and ANOVA for the correlation between lowest measured temperature at each station and respective number of days with T_max lower than 0 °C were calculated using statistical software R (R Core Team 2021).
DNA extraction and Sanger barcoding sequencing
DNA was extracted from the complete fruit fly specimens using a Qiagen Blood and Tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. For confirmation of the morphological identification results and for differentiation of the specimens from Germany compared to worldwide-acquired reference populations, COI was amplified by PCR and subsequently Sanger sequenced. Amplification of the barcoding region COI was conducted by PCR of undiluted DNA extracts using the insect-specific primers LCO1490 and HCO2198 of Folmer et al. (1994) and PCR conditions as published in EPPO PM7/129(2) (Anonymus Anonymous 2021). Prior to cycle sequencing using BigDye Terminator v3.1 (Applied Biosystems/Thermo scientific), PCR products were purified in an enzymatic approach. Purification was done using a mixture of Shrimp Alkaline Phosphatase (0.2 µl), Exonuclease I (0.2 µl) in molecular-grade water (1.6 µl) in a thermal protocol of 5 min at 37 °C for enzymatic digestion of remaining primer and polymerase stopped by 10 min incubation at 80 °C. Sanger sequencing electrophoresis and data sampling were performed using an automated sequencer (ABI PRISM 2100 Genetic Analyzer, Applied Biosystems).
COI phylogenetic evaluation
Sequence editing was examined using Sequencher software for windows vers. 5.0 (Gene Codes Corporation, Ann Arbor, MI, USA). Closest matches to all specimens were determined using BLASTN sequence similarity search tool in the National Centre for Biotechnology Information (NCBI) database. Reference sequences of specimens from Germany and other populations were aligned together with an outgroup of tephritid and other fruit fly families (Rhagoletis complete, Drosophila subobscura and Phaonia trimaculata) using the program MAFFT vers. 5.731 (Katoh et al. 2002). Phylogenetic relationships were tested by neighbour joining coupled with 1000 replicates of nonparametric bootstrapping using Kimura two-parameter (K2P) distances as implemented in software MEGA 7.0.26 (Kumar et al. 2016).
Population genetics: laboratory assay
DNA extracted for COI Sanger sequencing was subsequently used for microsatellite analysis to investigate population genetics based on 10 microsatellite (SSR) loci (Bonizzoni et al. 2000). Individual genotyping was conducted using three separate mixtures for three to four probes each containing primers in a concentration of 100 µM, 12.5 µl of GoTaq Colorless twofold concentrated mastermix (Promega, Madison/WI, USA) and 9.1 µl molecular-grade water. Three microlitres of DNA extract, fivefold diluted with molecular-grade water, was used as template for each PCR. Forward primers of each locus were individually labelled with a fluorescence dye and incorporated into the SSR fragments during PCR. Detection of the amplified labelled SSR fragments was conducted by capillary electrophoresis using an automated sequencer (ABI 3130 XL Genetic Analyzer, Applied Biosystems). Loci, mixture composition, labelling dye of the forward primer and primer concentration used are given in Table 1.
PCR of microsatellite loci was carried out as follows: two minutes of initial denaturation at 95 °C, 30 s at 95 °C, 30 s at 50 °C, 30 s at 72 °C for 35 cycles and 10-min final elongation at 72 °C.
Fragment length scoring was done using a size standard added to each mixture of labelled PCR fragments directly in the automated sequencer and corrected visually if necessary.
Population genetics: data analysis
Main parameters for the genetic variation of individuals within a geographically defined population are (i) the degree of polymorphism (P) within each locus of a single population; (ii) the mean number of alleles per locus (A) with emphasises on alleles unique to a single population (private alleles, AP), and (iii) the average number of (expected and observed) heterozygous individuals (H). Allele frequencies were used to estimate the degree of genetic identity (I), allele fixation indices (F) and to calculate genetic distances between populations (D; D = − logeI) (Milani et al. 1989). Prior to the analysis of genetic variation parameters, loci were tested for experimental bias as the presence of null alleles, allelic dropout and scoring error using MicroChecker 2.2.3 (van Oosterhout et al. 2004). Input files for subsequent analyses examined in Arlequin 3.5.1.3, Bottleneck 1.2.02 and MSA 4.05 were generated in Convert 1.31 (Glaubitz 2004). Pairwise linkage disequilibrium (LD) for unravelling erroneous correlations between two or more of the loci within each single geographically distinct population was analysed using Arlequin 3.5.1.3 (Excoffier et al. 2005) with Markov chain Monte Carlo (MCMC) parameters of 100,000 steps in 1000 dememorizations. Heterozygosity and therefore the conformity of each locus according to the hypothesis of the Hardy–Weinberg Equilibrium (HWE) were also calculated in Arlequin 3.5.1.3. Because multiple comparisons artificially inflate type I errors, a sequential Bonferroni correction (Rice 1989; Sokal and Rohlf 2012) was performed on P-values resulting from tests of HWE and pairwise linkage disequilibrium (LD) to determine significant differences of the experiment-wise type I error rate of 5%. To test for the hypothesis that habitats of C. capitata experienced historical bottlenecks (changes in population size) accompanied by an excess of heterozygosity in a number of specimens due to founder events, a one-tailed Wilcoxon rank test using the two-phase model (TPM; 70% SMM:30% IAM) was calculated in Bottleneck 1.2.02; (Cornuet and Luikart 1996).
The F statistics (Fst) of population structure (Wright 1951) is an important tool that measures the heterozygote deficit relative to its expectation under the Hardy–Weinberg Equilibrium. Fst can be interpreted as a measure of differentiation between geographically distanced populations and enables the quantification of these changes in allele frequency between such (sub-)populations in relation to the total population (Goudet 1993). Fst increases the more allele frequencies diverge between two sampled (sub-)populations. The fixation index or inbreeding coefficient Fis partitions the heterozygote deficit of individuals in relation to the (sub-)population. Negative values of Fis can therefore indicate a heterozygote excess while positive values show heterozygote deficits. Fst was calculated using the Micro Satellite Analyser (MSA, Dieringer and Schlötterer 2003). Calculations of Fis values and tests for their significance were determined using Fstat (Goudet 2003). Fst values between different populations of C. capitata were subsequently used for testing of isolation by distance. Isolation by distance (IBD, Wright 1943) is a hypotheses considering that individuals have been packaged and dispersed in discrete structures or clusters, respectively, with limitations in their continuous gene flow with increasing spatial distance. IBD was tested using a Mantel test as an in-between class analysis, which correlates with a matrix of Fst values with another matrix of natural logarithm (ln) of geographic distances between these populations. Mantel’s test was calculated using R-package ade4 (Thioulouse et al. 2018) in a Monte Carlo test using 999 permutations.
Allelic diversity (A), defined by the expected effective allele numbers compared to the exact numbers of private alleles per population (AP), was analysed using MSA and summarised for each sampling site per hand. Effective allele count (AE) was calculated as expected allele number in MSA using the SMM model under stepwise production of neutral alleles (Kimura and Ohta 1975). Genetic distances (D) between sample sites were calculated using Cavalli-Sforza and Edwards’ chord distances (Dc). Therefore, 5000 distance matrices were produced from resampled data (Micro-Satellite-Analyser) and analysed in Neighbor (phylip 3.68, Felsenstein 1989) to construct an unrooted majority-rule consensus neighbour-joining (NJ) tree with bootstrap values that was generated using R-package ape (Paradis and Schliep 2019). A Bayesian MCMC approach as employed in STRUCTURE 2.1 (Pritchard and Wen 2004) was used to detect the underlying genetic population among a set of individuals genotyped at multiple markers. STRUCTURE computes the proportion of the genome of each individual independent from geographical coordinates in a quantitative clustering method. In consequence, this software enables a qualitative view of population structure without geographical restrictions independent from coordinate data and changes in allele frequency across landscapes. The analysis aims to elucidate the smallest value of virtual populations K that captures the major structure in the data. The number of underlying K genetic clusters could therefore be calculated using the second-order rate of change of the likelihood (ΔK = m([L″K])/s[L(K)]) (Evanno et al. 2005). The plotted ΔK shows a clear peak at the point with the largest change of K indicating the most likely value of K. Based upon isolation and founder events, results of K values between K = 1 and K = 15 were evaluated using the likelihood values L(K) for 10 repeated runs per K, a burn-in period of 50,000 and MCMC lengths of 100,000. All calculations were performed using the admixture model as implemented in STRUCTURE to produce an estimate of K.
A principal component analysis (PCA), used to unravel changes in population structure between different years, was calculated in R-package gstudio (Dyer 2013) with graphical support of R-package ggplot2 (Wickham 2016).
Results
The official survey for the detection of C. capitata in Germany was conducted with slightly different objectives in the years 2015–2017. In 2015, the survey was focussed on apple orchards; in 2016, the focus had been extended to fruit trading places, stone fruit cultivation sites, and composting facilities. In 2017, the survey was continued on selected sites in a lower extent in some Federal States. In all years, medfly occurrence was surveyed from June to October. While in 2015 in total 15 specimens were trapped, in 2016 the number increased to 188 and decreased to 29 in 2017 (χ2 = 238.82; df = 2, P > 0.0001). Single specimens were trapped in most Federal States with clear emphasis in the southern parts (Baden-Wuerttemberg, Rhineland-Palatinate), and Brandenburg close to German-Polish-border in the north-eastern part of Germany. The temporal peak of fruit fly occurrence was detected in September correlating with ripeness of apple (Fig. 1). Although the traps were intended to lure male individuals by using trimedlure, a number of female specimens were trapped in all years. In years with low trapped individual numbers, female medflies were above average (2015: 9 female to 6 male specimens (χ2 = 0.6, df = 1, P = 0.43); 2017: 12–17; χ2 = 0.86, df = 1, P = 0.353) only in 2016 the male specimens could be more specifically lured (165 out of 188; χ2 = 1.5, df = 1, P = 0.22).
The lowest minimum temperature was measured at the meteorological station Dresden-Klotzsche on January 22nd in 2016 with − 13.3 °C within a period of 11 days with minimum temperatures below zero degree. The highest maximum temperature was found in Mannheim on 7 August 2015 measuring 39.7 °C. The lowest daily maximum temperature was found in Manchnow on January 3rd with − 8.9 °C within a period of six days with maximum temperatures not exceeding the zero degree mark. The number of days with maximum temperatures below 0 °C was lowest in 2015 with a single day in Mannheim and highest in 2017 with in total 21 days in Dresden–Klotzsche (see Supplementary Fig. 1a for all temperature profiles). Spearman rank correlation analysis revealed highest correlation between the number of days with Tmax < 0 °C and the lowest maxim temperature measured on each station (ANOVA P < 0.0001). Statistical differences between the number of days with maximum temperatures lower than 0 °C were found between 2015 and the years 2016 (W = 3; P = 0.019) and 2017 (W = 0; P = 0.005). Between 2016 and 2017 as well as between the different meteorological stations, no differences were observed.
For molecular investigation, only fruit fly sampled from 2016 and 2017 in Germany was available. Therefore, 176 out of 206 specimens available could be analysed successfully (2016:154; 2017:22). All of the molecular analysable specimens could be identified as individuals of C. capitata and used within both assays, phylogenetic analysis of COI Sanger sequencing data and microsatellite analysis. In total, 496 specimens from 11 countries (Table 2) were further analysed to determine their relationship with the German specimens. Most of the specimens from reference populations of other countries were collected in 2016. Samples from 9 out of 11 reference populations were taken from consignments of fruits imported to Germany and France. Most important host species of these populations were citrus fruits, such as Citrus sinensis for fruit flies with origin from South Africa, Tunisia and Egypt, as well as Psidium guajava (Brazil, Egypt), Citrus aurantiacum (Italy), pepper (Capsicum sp. from Togo, Cameroun) and Apple fruit (Lebanon, France). Almost all individuals of these international populations were analysable within COI Sanger barcoding sequencing and SSR with one exception. Nine DNA extracts from specimens of South-African medflies were successfully Sanger sequenced for COI but only five of them could be examined further in SSR analysis.
Based on 658-bp fragments of COI Sanger sequences, seven clades could be identified for the examined C. capitata specimens from German and worldwide origin (Fig. 2). German specimens were situated completely in clade 1 together with specimens from Egypt, Croatia, France, Italy and Tunisia sharing exactly the same COI sequence. Clade 2 comprised all specimens from Mediterranean origin. Except from a single sequence from a South-African medfly population distinguished by two base pairs difference to the specimens in clade 1, all African populations were completely separated from the remaining populations in Mediterranean area in clade 2 and the populations from Central Europe in clade 1. Medflies from African origin including these originating from Brazil were situated in the monophyletic clades 3–7.
Neighbour-joining phylogenetic tree (phylogram) based on K2P distance analyses of the mitochondrial COI region of 108 representative sequences of populations of C. capitata sampled in 12 countries. Clades were separated by tree architecture on branches with Bootstrap values exceeding 50% (from 1000 resamplings). Sequence numbers are given in brackets for countries of origin of the specimens. Different colours indicate the assignment of the respective specimens to the main virtual population of the country of origin as generated in software STRUCTURE (see Fig. 6b)
Laboratory preparation of SSR fragments was in most cases hampered by high DNA concentrations; therefore, DNA was up to fivefold diluted, and at least three times replicated in the case of negative PCR amplification (Eurofins, Ebersberg, Germany). Due to non-amplifiability in primer mixtures (see Table 1), PCR of loci Ccmic9 and Ccmic12 was carried out in single-plex reactions. This allows to amplify most of Ccmic9 (98.9%) but only 25.1% of Ccmic12. The latter clearly indicates the presence of null alleles, which could be confirmed in Micro-Checker analysis for Ccmic12 in populations of C. capitata from Germany and most of the other countries. Null alleles indicate events of non-amplification of present fragments from single allele loci and could be detected via homozygote excess. Null alleles were further observed for loci Ccmic4 and Ccmic8 (for both loci only German specimens), Ccmic6 (Brazil) and Ccmic9 (Togo). Lack of polymorphism occurred especially in Mediterranean populations of C. capitata (Italy, Tunisia, Lebanon, and Egypt) for two to three loci (Ccmic4, 7, 12) sharing the same allele in all genotypes. A number of scoring errors caused by single-nucleotide base drop out was obvious in locus Ccmic12 in French population and Ccmic6 in fruit flies from Brazil. Linkage disequilibrium (LD) was detected for a range of pairwise comparisons between loci in several populations of C. capitata. German populations of the medfly showed in total 21 significant deviations from linkage equilibrium mainly concerning loci Ccmic15, 8, 7, 13 and 6. Nearly the same situation could be observed in France. Maximum of LD was found in specimens sampled in Spain with in total 33 pairwise deviations from linkage equilibrium. All of the remaining populations showed no LD. LD is an indicator for non-random low numbers of genetic recombination events due to closely linked pairs of loci e.g. on the same chromosome. Populations distant from the place of species origin arose from a subset of genomes, resulting in a bottleneck in population size in which certain combinations of alleles were overrepresented leading to the increased LD and possible deviations from HWE (Hartl 2020). However, German and French populations showed no deviations from Hardy–Weinberg equilibrium. Completely unexpected for the high amount of LD is the heterozygote excess in the Spanish population resulting from a high discrepancy between observed and expected heterozygosity. This is furthermore manifested by the highest negative inbreeding coefficient (Fis) and a moderate significant bottleneck detected there (Table 2).
Detected number of alleles and private alleles (Table 2; Fig. 3) largely depends on the sampling design and the number of specimens sampled in an actual population. Due to the intensive surveys in Germany and France, as expected, the highest allele numbers were detected in the French populations due to the accumulation of alleles. Allelic diversity was 34.7 ± 14.9. The high standard deviation reflected the huge differences in populations due to different sample sizes. However, the highest effective allele numbers normalised for the number of sampled specimens were found in medfly populations of Togo and the Republic of South Africa, followed by the populations in Spain, France and Cameroon. The lowest allelic diversity was detected in populations from countries in the eastern Mediterranean area. The number of private alleles, which only appear once in single populations exclusively, was also highest in the populations sampled in the African countries and France. Looking on allele fragment length (data not shown), the probably most recent populations found in Germany and France showed fragments only present in the African populations close to species origin mixed with such of the derived populations from the Mediterranean basin. The Spanish populations were surprisingly much closer related to the African populations than to the Mediterranean ones regarding their composition of detected SSR-fragments. The German populations were therefore in between the French and Croatian populations.
Observed (Hobs) as well as expected heterozygosity (Hexp) decreased with geographic distance and stage of derivation of the sampled individuals beginning from (i) close to the origin of C. capitata in Africa (Togo, Cameroon, and South Africa) to (ii) the Southern American populations (Brazil), (iii) African Mediterranean outbreaks (Tunisia, Egypt), (iv) outbreaks in European Mediterranean areas (Italy, France, Croatia) and (v) from Central Europe (Germany) with the exception of Spanish populations. Surprisingly heterozygosity was highest in Spain. The lowest heterozygosity was found in Lebanon in the east of the Mediterranean basin (Fig. 4a). With the exception of individuals collected from Spain, all populations were in Hardy–Weinberg equilibrium. Ccmic12 was the only locus not in HWE within most of the populations due to the high numbers of null alleles.
Pairwise Fst values (Table 3) varied widely between 0.035 (populations from France/Croatia) and 0.49 (Cameroon/Lebanon) with an overall mean Fst = 0.25 ± 0.11. Almost all Fst values differ significantly from zero in order to the heterogeneity Chi-square test. Again, the largest differences occurred between sub-Saharan populations and Oriental-Mediterranean populations of C. capitata. The effect of spatial scale on gene frequencies for specimens trapped in Germany was lowest in comparison with populations sampled in France (0.04), followed from these sampled in Croatia (0.09), and Egypt (0.12) when spatial scale was partitioned into 12 geographic groups for countries medflies derived from. To test for the hypothesis of isolation by distance (Wright 1943), Mantel’s test revealed significant correlation between Fst values on the one hand and spatial distances on the other hand (P < 0.01) even in the case of lower geographic resolution when capitals were representatively set for the mostly unknown sites of fruit consignment origin in the different reference populations. A linear model calculated for Fst values and log natural geographic distances of all 66 pairwise comparisons confirmed these results (R2 = 0.35; P < 0.001) (Fig. 4b). The results in both statistical analyses showed that isolation by distance shapes the contemporary present populations of C. capitata. Positive Fis values occurred in the populations sampled in Germany, Croatia, France, Lebanon as well as in South Africa and Togo (Table 2).
Unrooted neighbour-joining trees using Cavalli-Svorza and Edwards’ (1967) chord distance (DC). Bootstrap values at each node were calculated using a consensus tree derived from 5000 distance matrices. Nodes supported by ≥ 50% bootstrapped values are included. Figure 4a shows populations of 12 countries were C. capitata was sampled, and Fig. 4b depicts populations sampled in three departments in France and nine Federal States of Germany. Numbers of sampled specimens per population and time of sampling are given in brackets
Different phases of worldwide spread of C. capitata into suitable subtropical and tropical habitats as reported in literature (Enkerlin et al. 1989; Fimiani 1989; Hancock 1989) (a). STRUCTURE analysis of genotypes of 672 medfly specimens (b) represented by a bar partitioned into K = 3 segments representing the specimen’s estimated membership fractions calculated by STRUCTURE for each of the three clusters
Focussed on the populations sampled in Germany, the largest differences of Cavalli-Sforza & Edwards’ genetic distance (DC) were observed in relation to the specimens sampled in Cameroon (0.68) followed by those of the remaining African populations. The closest relation was found between the German and French populations (DC = 0.16). The NJ tree (Fig. 5a) based on DC distinguished populations sampled in Africa from those in Germany and France on a second clade of the tree and the populations sampled in the Mediterranean area positioned in a third clade. While the populations of Brazil and Spain were more related to the African populations, the Croatian population connected the French and German populations to the Mediterranean ones.
Runs in STRUCTURE software elucidated a peak of change for second-order rate of log likelihood over 10 runs at K = 3 clusters (Supplemental Fig. 2) with a mean log likelihood L(K = 3) of -11,923. These clusters were (1) African populations (80–98%), including populations from Brazil (79%) and Spain (80%); (2) Mediterranean populations comprising populations from Croatia (77%), Egypt (80%), Tunisia (92%), Lebanon (93%), and the main part of France (66%), as well as 32% of German populations; (3) Germany (66%) and 27% of specimens sampled in France (Fig. 6b). Remarkable portions of 13 and 16% with one and four specimens of the populations sampled in Croatia and Egypt, respectively, could also be assigned to the third cluster.
As already indicated by population genetic descriptors (F-statistics, A, D, P, H) presented before, results generated in STRUCTURE demonstrated most clearly that a third wave of most recent spread of the medfly now is manifested also in new pattern formations of genetic variability. Starting from cluster 1 (C1) mainly represents specimens from population close to the origin and from direct introductions to South America and Spain up to the derived Mediterranean cluster 2 (C2) and cluster 3 (C3) corresponding to most recent outbreaks. A significant decrease was found between cluster 1 and the remaining clusters in allele count (AN, AP and AE) and heterozygosity but there was no difference between cluster 2 and cluster 3 (Table 4).
Fst showed a closer relation between clusters 1 and 2 (0.1293) than between both clusters and cluster 3, which is intermediate between the specimens from Mediterranean basin and the sub-Saharan specimens with Fst values approximate to 0.2 in relation to each of the other clusters (Table 4). For Fis, we found a hierarchical relationship from negative Fis observed in cluster 1, to a moderate deficit in heterozygosity in the remaining clusters higher in cluster 3 than in cluster 2 (Table 2).
In conclusion, while outbreaks at the beginning of the twentieth century could be rather attributed to single source populations as shown in South America, the most recent outbreaks in Germany and France built their own population stage. Cluster 3 is distinguished from the African and Mediterranean ones by new occurring SSR fragments as shown by seven private alleles in French populations (Table 2) as well as absorbing features in terms of allele composition from all existing C. capitata populations. Several population genetic effects potentially form these most derived populations as there are (i) local admixture of specimens from different origin, (ii) strong shifts in population size (bottleneck) due to local food availability and climate conditions accompanied by (iii) gene drift, which is typically observed as random change in allele-frequency especially occurring within small populations (Weir 1996).
The question, whether overwintering of C. capitata in Germany between the years 2016 and 2017 took place, is more difficult to answer than the question of potential descent. Because low numbers of individuals from small outbreak populations in local orchards were trapped, comparable individual numbers higher than ten individuals for each year of monitoring could only be achieved by aggregating several of these populations from the same German Federal State and Department in France. Only in the case of medfly populations from Baden-Wuerttemberg, populations comparable in size and geographic area were sampled in two years, 2016 and 2017. These populations could be distinguished by means of genetic distance between the different years (Fig. 5b). Individual specimens genetic comparison as analysed in a principal component analysis (PCA, Supplementary Fig. 3) revealed larger differences for specimens captured in 2016 compared to these sampled in 2017. Higher genetic variability in 2016 was expressed by broader dispersal of specimens points in PCA largely but not completely overlapping with the genetic variability detected in 2017. However, the shift in population genetic variability between different years is difficult to resolve from natural occurring intra-population events as gene-drift and bottlenecks which are found in not completely admixed populations even when using SSR-marker.
At one sampling site situated near Frankfurt/Oder, overwintering of a single population from 2015 to 2016 is very likely, as in 2015 one specimen was detected in a trap closely situated to the trap in which in 2016 numerous specimens were detected. The traps were located in a plum orchard without application of plant protection measures where fruits are no longer harvested but allowed to rot on the ground, with sweet cherry and apple cultivated in direct vicinity of the site. In total, 119 predominantly male specimens were trapped there between August and October 2016. However, as the specimens caught there in 2015 were not available for the respective examinations, a direct lineage could not be proven. In 2017, no further specimens could be detected on this site.
Discussion
Worldwide trade of plants and plant-derived commodities, such as fruits, seeds and wood, can contribute to the dissemination of numerous organisms that might be more or less closely attached to the imported consignments. Early detection and control is the basis to prevent high economical, ecological or social impacts on natural and agricultural habitats. Surveillance can provide information to the responsible National Plant Protection Organisations (NPPOs) and can be an important technical basis for many phytosanitary measures, e.g. issuing of phytosanitary export certificates, establishment of pest free areas where the organism is not yet present, pest reporting as well as determination of the pest status in an area (FAO 2018). The International Plant Protection Convention (IPPC) established a concept for the official determination of the status of a specific pest within a defined area in relation to its occurrence and distribution (FAO 2017). According to this standard, pest status is described under three main categories as present, absent or transient. If sufficient information is available, the status can be further characterised by added phrases, such as ‘present: only in some areas’ or ‘absent: pest eradicated’. The category transient should be used, if a pest is present but establishment is not expected to occur based on technical evaluation, e.g. lack of sufficient host plants or unsuitable climatic conditions.
Although C. capitata is present in the European Union since at least 1842 (Malaga, Spain Fimiani 1989) and therefore well established in countries around the Mediterranean basin, populations of the organism were so far not established in Middle European countries (Fischer-Colbrie and Busch-Petersen 1989). The species northern distribution is currently restricted to the 41th parallel north (Robinson and Hooper 1989; Israely et al. 2004), as suitable habitats for establishment should possess mean annual temperatures not lower than 10 °C (Mwatawala et al. 2015). Nonetheless, C. capitata has been observed in Germany several times since 1934 (Fischer-Colbrie and Busch-Petersen 1989) as it is often connected to non-regulated fruit imports from EU member states where it is established. Regarding climate change and increasingly mild winter temperatures in some areas, the question was, if establishment of single populations of fruit flies in Germany could have already been occurred.
The presented results from the medfly survey conducted between 2015 and 2017 show only low abundances of C. capitata in relation to pome fruits in most parts of Germany with main focus on the southern parts (Federal State Baden-Wuerttemberg) and only single specimens trapped further north. An exception was a spot in the wider vicinity of Berlin (near Frankfurt-Oder) where in 2016 a maximum of 124 specimens could be trapped in an abandoned stone fruit orchard.
Tracing back the origin of specimens found in Germany in 2016 and 2017 by different genetic analysis showed that they could be clearly assigned to populations in one or several countries where the fruit fly is known to be well established. Although the selected specimens analysed for this study represent only a small part of C. capitata populations present worldwide that have derived from just a small number of plant hosts the findings of this study support the hypothesis that the populations of the medfly in Germany are mainly transient. COI phylogeny of C. capitata already reflected the main aspects of medfly dispersion as reported in the literature (e.g. Malacrida et al. 1998; Fimiani 1989), with highest differences between German populations and populations sampled in African countries. However, genetic distance between the most distantly related medflies in clade 2 and these in clades 5–7 was low with 0.0123. COI resolution depends on in total 11 informative base characters out of 658 bp. As expected, COI phylogenetic analysis therefore enables only a restricted differentiation in terms of identification of the origin of German medfly specimens, but allowed a first assignment of these specimens together with populations present in France and eastern Mediterranean areas.
The use of appropriate primers for the amplification of 10 small simple repeat microsatellite loci from literature published by Bonizzoni et al. (2000) showed a couple of limitations and deficiencies for several loci. Observed limitations in our study were (i) more than 1000 null alleles with most of them on locus Ccmic12, (ii) low locus polymorphisms at least in some Mediterranean populations, (iii) homozygote excess and (iv) scoring errors by nucleotide base drop out. Bonizzoni et al. (2000) already described a lack of heterozygotes in Ccmic12 in a single population from Madeira showing no heterozygote specimens at all and the authors assumed further that their observed excess of homozygotes in African populations could be probably caused by null alleles. In addition, scoring errors were found in our analysis for Ccmic6 in samplings from Brazil and Ccmic12 in fruit flies from France. This is in agreement with Bonizzoni et al. (2000), who already reported uneven fragment length to occur naturally in very low frequencies, suggesting a mutation process caused by either strand slippage or point mutations. Lack of polymorphism could be due to small sampled population sizes but also to low genetic variation. The increasing number of such limitations of the SSR assay may reflect the fast evolution process of the medfly undergoing numerous bottlenecks and in its consequence several founder effects. Severe population bottlenecks often occur in nature when a small group of disseminated individuals from an established subpopulation founds a new geographically isolated subpopulation. The random genetic drift and the loss of genetic variation that occurs when a new population is established by a very small number of individuals from a larger population accompanying such a founder event is defined as founder effect (Hartl 2020). Some populations of C. capitata experienced several founder effects during their extensive spread since the molecular tools had been established 20 years ago what explains the current performance of the SSR assay.
By using SSR loci qualitative allelic diversity (A), heterozygosity (H), fixation indices (F) and genetic distance (DC) showed that populations from France and Croatia strongly resemble those collected in Germany but are not identical with these. With no consideration of the temporal and geographical relations, using the Bayesian admixture approach as implemented in STRUCTURE the different German populations of C. capitata are furthermore not homogenous in its descent (Fig. 6). In relation to available reference specimens, German populations are most concordant with those sampled in France 2016 but also influenced by populations from the Mediterranean area (Croatia, Egypt, but not the specimens sampled from Italy, see Table 3 for Fst values). Populations found in Germany did not completely resemble any of the reference populations. Spanish population vouchers were shown to influence population genetic structure of German populations only marginally.
The genetic variance observed in our study between medfly populations of different countries almost resembles that from earlier investigations of the fruit fly in the 1990s (e.g. Malacrida et al. 1992, 1998 for results of alloenzyme variation analysis). Observed heterozygosity (Hobs) as well as expected heterozygosity (Hexp) decreased with geographic distance to species origin. The same was found for allelic diversity decreasing in correlation with geographic distance, date of introduction of the medfly and stage of derivation of the more distant populations from these present at the species origin (Malacrida et al. 1992, 1998; Bonizzoni et al. 2000). The only exception from earlier findings was a de novo increased observed heterozygosity in Spain and France indicating recent new introduction and establishment of Mediterranean fruit fly populations in these countries. As confirmed in literature (Malacrida et al. 1998), almost all Fst values differ significantly from zero shown by heterogeneity chi-square test but the results in our analysis clearly confirmed the hypothesis of populations isolation by distance (Wright 1943; Fig. 4). This finding is in contrast to the results from Malacrida et al. (1998) who showed that spatial distance was separated into nine geographic groups representing 17 populations from 12 different countries. The authors found significant patterns of isolation by distance only in regression analysis when most recent outbreak areas of C. capitata (Latin America, Pacific and Australia) were removed from the analysis. Positive findings of the inbreeding coefficient Fis significantly different from zero inter alia in German populations. Fis indicates possible increased co-ancestry between offspring and is therefore characteristic of small restricted populations present on isolated local sites (Goudet 1993). The presence of small distinguishable populations in Germany indicates that low numbers of specimens might survive for some time in sheltered places, allowing C. capitata to reinvade new regions when seasonal weather conditions become more suitable in spring and summer (Carey 1991). However, the results do not indicate the presence of established populations in Germany but strongly support the hypothesis that the medfly populations in Germany are still transient. In this case, specimens were repeatedly introduced within the last years by the import of fruits. Furthermore, within the same geographic scale it is possible to distinguish between the populations sampled in 2016 and 2017 (Fig. 5b) although there existed an overlap (Supplementary Fig. 3), which is probably explainable by a shift due to a number of generations and host alteration in between. Re-invasion rather than overwintering of C. capitata in 2017 could be further explained by different temperature conditions observed in the years the monitoring was conducted from 2015 to 2017. While in 2015, only 1–5 frost days with a maximum temperature lower than zero degree were found in 2016 and 2017 the number increased to 13 and 21 days at the meteorological station in Dresden-Klotzsche. Nevertheless, climate conditions are unevenly distributed in Germany, which could be shown for Konstanz where 3 frost days appeared in 2016 as compared to Manchnow close to Frankfurt/Oder with 14 frost days in the same year. These could further explain the decline of the population at the most infested sampling site situated near Frankfurt/Oder between 2016 and 2017 as stated above. Besides the pattern of different origins of the populations collected in Germany, a high percentage of females was captured especially in 2015 and 2017. This indicates the absence of males as a typical characteristic of small populations where unmated mature females were attracted to sexual male-attractant para-pheromones as trimedlure and medlure (Nakagawa et al. 1970).
However, this situation might change in the future, as shown by a recent study from Gilioli et al. (2021) who presented results from a nonlinear modelling approach of C. capitata on the basis of recent (2020) and future climate scenarios (2030 and 2050). They elucidated temperature as the main factor driving the establishment of the medfly also in areas more northern than the 41st parallel in Europe. Further consideration of differences between larval and adult population abundance reflecting intra-specific competition as a density-dependent factor within the nonlinear model, predicts a shift of the dispersal of C. capitata up to the 46th parallel north in continental climate in the North of Italy and even to the 48th parallel north in oceanic climate in France. Due to an expected adaptation of the northern populations to changed climate conditions (Gilioli et al. 2021) it is already visible in our STRUCTURE analysis data that cluster 3 contains a potentially adapted dispersal stage of C. capitata mainly present in France and Germany (Fig. 6) with lowest genetic variation (Tables 2 and 4). For Germany it could be expected that the areas with higher abundance of the medfly elucidated in the surveys conducted between 2015 and 2017 will be the ones were the fruit fly first becomes established. Although C. capitata is not regulated as a quarantine pest organism, it is expected to potentially damage pome and stone fruit orchards to a comparable extent as Drosophila suzukii in wine and cherry orchards (Baufeld et al. 2010). Additionally, C. capitata can act as a temperature threshold indicator for establishment of other important pests, especially tropical fruit flies like Ceratitis rosa, Bactrocera dorsalis or Bactrocera zonale, which are regulated as Union quarantine pests in Implementing Decision (EU) 2019/2072 (European Commission 2019) and the establishment of which can pose a great risk to fruit growers. Continuous plant pest surveys on C. capitata can therefore be an important part of the national early warning system to detect the climatic threshold for establishment of regulated union quarantine pests in time and to prevent considerable damage on cultivation sites as well as in natural habitats.
References
Anonymous (2021) PM 7/129 (2) DNA barcoding as an identification tool for a number of regulated pests. EPPO Bull 51:100–143. https://doi.org/10.1111/epp.12724
Barr NB (2009) Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J Econ Entomol 102:401–411. https://doi.org/10.1603/029.102.0153
Baufeld P, Schrader G, Unger JG (2010) The Cherry vinegar fly—Drosophila suzukii—an emerging risk for fruit and wine growing. J Kult 62:183–186
Bonizzoni M, Malacrida AR, Guglielmino CR, Gomulski LM, Gasperi G, Zheng L (2000) Microsatellite polymorphism in the Mediterranean fruit fly, Ceratitis capitata. Insect Mol Biol 9:251–261. https://doi.org/10.1046/j.1365-2583.2000.00184.x
CABI/EPPO (1997) Quarantine pests of Europe. CAB Intern, S.l.
Carey JR (1991) Establishment of the Mediterranean fruit fly in California. Science 253:1369–1373. https://doi.org/10.1126/science.1896848
Cayol JP, Causse R (1993) Mediterranean fruit fly Ceratitis capitata Wiedemann (Dipt., Trypetidae) back in Southern France. J Appl Entomol 116:94–100. https://doi.org/10.1111/j.1439-0418.1993.tb01172.x
EU Commission (2019) COMMISSION IMPLEMENTING REGULATION (EU) 2019/2072
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014. https://doi.org/10.1093/genetics/144.4.2001
Dieringer D, Schlötterer C (2003) microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169. https://doi.org/10.1046/j.1471-8286.2003.00351.x
Dyer RJ (2013) gstudio: Spatial utility functions from the Dyer laboratory. https://CRAN.R-project.org/package=gstudio
Enkerlin D, Garcia LR, Lopez FM (1989) Pest status: Mexico, Central and South America. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3A, pp 83–90
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform. 1:117693430500100. https://doi.org/10.1177/117693430500100003
FAO (2017) ISPM 8 Determination of pest status in an area
FAO (2018) ISPM 6 Surveillance
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166
Fimiani P (1989) Pest status - Mediterranean Region. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3A, pp 39–50
Fischer-Colbrie P, Busch-Petersen E (1989) Pest status: Temperate Europe and West Asia. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3A, pp 91–99
Fletcher BS (1989) Movement of tephritid fruit flies. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3B, pp 209–217
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Gilioli G, Sperandio G, Colturato M, Pasquali S, Gervasio P, Wilstermann A, Dominic AR, Schrader G (2021) Non-linear physiological responses to climate change: the case of Ceratitis capitata distribution and abundance in Europe. Biol Invasions. https://doi.org/10.1007/s10530-021-02639-9
Glaubitz JC (2004) Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310. https://doi.org/10.1111/j.1471-8286.2004.00597.x
Goudet J (1993) The genetics of geographical structured populations. PhD thesis, Bangor, Gwynedd, Wales UK
Goudet J (2003) Fstat (ver. 2.9.4), a program to estimate and test population genetics parameters. Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet [1995]
Hancock DL (1989) Pest status - Southern Africa. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3A, pp 51–58
Harrell Jr FE (2021) Hmisc: Harrell Miscellaneous. https://CRAN.R-project.org/package=Hmisc
Hartl DL (2020) A primer of population genetics and genomics, 4th edn. Oxford University Press, Oxford
Israely N, Ritte U, Oman SD (2004) Inability of Ceratitis capitata (Diptera: Tephritidae) to overwinter in the Judean hills. J Econ Entomol 97:33–42. https://doi.org/10.1093/jee/97.1.33
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Kimura M, Ohta T (1975) Distribution of allelic frequencies in a finite population under stepwise production of neutral alleles. Proc Natl Acad Sci U S A 72:2761–2764. https://doi.org/10.1073/pnas.72.7.2761
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Malacrida AR, Guglielmino CR, Gasperi G, Baruffi L, Milani R (1992) Spatial and temporal differentiation in colonizing populations of Ceratitis capitata. Heredity 69:101–111. https://doi.org/10.1038/hdy.1992.102
Malacrida AR, Marinoni F, Torti C, Gomulski LM, Sebastiani F, Bonvicini C, Gasperi G, Guglielmino CR (1998) Genetic aspects of the worldwide colonization process of Ceratitis capitata. J Hered 89:501–507. https://doi.org/10.1093/jhered/89.6.501
Meats A, Smallridge CJ (2007) Short- and long-range dispersal of medfly, Ceratitis capitata (Dipt., Tephritidae), and its invasive potential. J Appl Entomol 131:518–523. https://doi.org/10.1111/j.1439-0418.2007.01168.x
Milani R, Gasperi G, Malacrida AR (1989) Ceratitis capitata genetics: biochemical genetics. In: Robinson AS, Hooper G (ed) Fruit flies: their biology, natural enemies and control, 3B, pp 33–54
Morales P, Cermeli M, Godoy F, Salas B (2004) A list of Mediterranean fruit fly Ceratitis capitata Wiedemann (Diptera: Tephritidae) host plants based on the records of INIA-CENIAP museum of insects of agricultural interest. Entomotropica 19:51–54
Mwatawala M, Virgilio M, Joseph J, Meyer M de (2015) Niche partitioning among two Ceratitis rosa morphotypes and other Ceratitis pest species (Diptera, Tephritidae) along an altitudinal transect in Central Tanzania. Zookeys. https://doi.org/10.3897/zookeys.540.6016
Nakagawa S, Farias GJ, Steiner LF (1970) Response of female mediterranean fruit flies1 to male lures in the relative absence of males. J Econ Entomol 63:227–229. https://doi.org/10.1093/jee/63.1.227
Paradis E, Schliep K (2019) ape 5.0: an environment for modern phylogenetics and evolutionary analysis in “R.” Bioinformatics 35:526–528
Pritchard JK, Wen W (2004) Documentation for STRUCTURE software, vers. 2
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225. https://doi.org/10.1111/j.1558-5646.1989.tb04220.x
Rigamonti IE (2004) Contributions to the knowledge of Ceratitis capitata Wied. (Diptera, Tephritidae) in Northern Italy: II. Overwintering in Lombardy. Boll Zool Agr Bachic 36:101–116
Sokal RR, Rohlf FJ (2012) Biometry: the principles and practice of statistics in biological research, 4th edn. Freeman, New York
Thioulouse J, Dray S, Dufour A, Siberchicot A, Jombart T, Pavoine S (2018) Multivariate analysis of ecological data with ade4. Springer. https://doi.org/10.1007/978-1-4939-8850-1
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Weir BS (1996) Genetic data analysis: methods for discrete population genetic data, 2nd edn. Sinauer, Sunderland
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer, New York
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354. https://doi.org/10.1111/j.1469-1809.1949.tb02451.x
Acknowledgements
The authors of this report thank the EUPHRESCO initiative for supporting this research. We thank Jennifer Elsner and Dr. Helena Ziert (Eurofins Medigenomix GmbH, Ebersberg, Germany) for execution of laboratory work, SSR-fragment scoring, and successful trouble shooting in all practical aspects, and the National Plant Protection Organisations of the Federal States of Germany for conduction of the surveys on C. capitata. We thank Valérie Balmès from ANSES Plant Health Laboratory (Rennes, France), Nuria Sierras, the head of Plant Health R&D at Bioiberica (Barcelona, Spain), and Dr. Mario Bjeliŝ (University of Split, Croatia) for the provision of European and worldwide reference populations of C. capitata. The authors would like to thank an anonymous reviewer for giving valuable advice on further data interpretation, and Dr. Kathleen Gärtner for her help to improve the English language in the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Funding was provided by National Outstanding Youth Science Fund Project of National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical responsibilities
The authors declare that (i) the manuscript has not been submitted to another journal simultaneously, (ii) the submitted work contains original data not published before, (iii) the data published are not split up into several parts to increase the personal number of publications, (iv) the current publication is not a secondary publication with the same content, (v) the presented data were not manipulated, and (vi) no data, text or theories by others not explicitly cited for their origin are presented in the current publication.
Clarification of authorship
All authors agree with the content of this publication and consent to submit this article to the Journal of Plant Diseases and Protection. The authors declare that they obtained content from the responsible authorities of the Julius Kühn-Institute for the article. The authors declare that they have no conflict of interest with the data published here. All authors whose names appear on this submission made substantial contributions to the conception or design of the work, or the acquisition of specimen material of C. capitata, or interpretation of data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
König, S., Steinmöller, S. & Baufeld, P. Origin and potential for overwintering of Ceratitis capitata (Wiedemann) captured in an official survey in Germany. J Plant Dis Prot 129, 1201–1215 (2022). https://doi.org/10.1007/s41348-022-00605-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-022-00605-8