Abstract
Purpose of Review
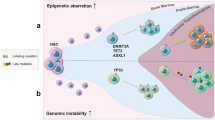
Advances in high-throughput methods have enabled the molecular characterization of leukemias and have improved our understanding of their clonal evolution from leukemogenesis in hematopoietic stem/progenitor cells to overt diagnosable disease.
Recent Findings
It has now been revealed that during leukemia’s development and progression, genetic alterations accumulate according to the principles of Darwinian evolution. Drug resistance often emerges from changes in evolutionary trajectories of disease through selection of subpopulations that have greater fitness under therapy. In this manuscript, we will review recent data on prevalence of highly branched evolutionary patterns in myeloid and lymphoid leukemias and discuss how different treatment strategies differentially shape leukemia’s clonal architecture.
Summary
Increasing evidence on clinical impact of small pre-malignant clones prior to diagnosis and small resistant clones during treatment strongly suggests that highly sensitive experimental and mathematical models are necessary for accurate dissection of hematopoietic populations and robust identification of predictive markers for disease transformation and relapse.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–81.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98.
Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355–61.
Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484.
• Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–21 A large-scale genomic study of more than 1500 AML patients showing prognostic power of genomic classification.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–33.
Mossner M, Jann JC, Wittig J, Nolte F, Fey S, Nowak V, et al. Mutational hierarchies in myelodysplastic syndromes dynamically adapt and evolve upon therapy response and failure. Blood. 2016;128(9):1246–59.
•• Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018 One of the first studies to directly link CHIP to development of AML.
•• Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nature Medicine. 2018;24(7):1015–23 One of the first studies to directly link CHIP to development of AML.
• Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157–70 A comprehensive review on CHIP and its evolution to hematological malignancies.
Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017.
Severson EA, Riedlinger GM, Connelly CF, Vergilio JA, Goldfinger M, Ramkissoon S, et al. Detection of clonal hematopoiesis of indeterminate potential in clinical sequencing of solid tumor specimens. Blood. 2018;131(22):2501–5.
Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8.
Arends CM, Galan-Sousa J, Hoyer K, Chan W, Jager M. Yoshida K, et al. Leukemia: Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis; 2018.
Tiacci E, Pettirossi V, Schiavoni G, Falini B. Genomics of hairy cell leukemia. J Clin Oncol. 2017;35(9):1002–10.
Chung SS, Kim E, Park JH, Chung YR, Lito P, Teruya-Feldstein J, et al. Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med. 2014;6(238):238ra71.
Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4(9):1088–101.
Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20(2):246–59.
Marsilio S, Khiabanian H, Fabbri G, Vergani S, Scuoppo C, Montserrat E, et al. Somatic CLL mutations occur at multiple distinct hematopoietic maturation stages: documentation and cautionary note regarding cell fraction purity. Leukemia. 2018;32(4):1041–4.
Alpar D, Wren D, Ermini L, Mansur MB, van Delft FW, Bateman CM, et al. Clonal origins of ETV6-RUNX1(+) acute lymphoblastic leukemia: studies in monozygotic twins. Leukemia. 2015;29(4):839–46.
Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319(5861):336–9.
Sanjuan-Pla A, Bueno C, Prieto C, Acha P, Stam RW, Marschalek R, et al. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood. 2015;126(25):2676–85.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–63.
Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111(7):2548–53.
Heath EM, Chan SM, Minden MD, Murphy T, Shlush LI, Schimmer AD. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31(4):798–807.
Gerstung M, Papaemmanuil E, Martincorena I, Bullinger L, Gaidzik VI, Paschka P, et al. Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet. 2017;49(3):332–40.
• Ferrando AA, Lopez-Otin C. Clonal evolution in leukemia. Nat Med. 2017;23(10):1135–45 A comprehensive review of clonal evolution in acute and chronic leukemias.
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–26.
Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–30.
• Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(7):601–12 This study showed the differential dynamics of TET2-first-mutated vs. JAK2-first-mutated myeloproliferative neoplasms.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–27 quiz 99.
Wang L, Fan J, Francis JM, Georghiou G, Hergert S, Li S, et al. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res. 2017;27(8):1300–11.
Wang J, Khiabanian H, Rossi D, Fabbri G, Gattei V, Forconi F, et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. Elife. 2014;3.
Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129(12):1595–606.
Cortes J, Jabbour E, Kantarjian H, Yin CC, Shan J, O'Brien S, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110(12):4005–11.
Falini B, Martelli MP, Tiacci E. BRAF V600E mutation in hairy cell leukemia: from bench to bedside. Blood. 2016;128(15):1918–27.
Tiacci E, Park JH, De Carolis L, Chung SS, Broccoli A, Scott S, et al. Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. N Engl J Med. 2015;373(18):1733–47.
Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122–30.
• Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Fama R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–47 The first systematic study to uncover the impact of small TP53-mutated clones on CLL prognosis.
Rasi S, Khiabanian H, Ciardullo C, Terzi-di-Bergamo L, Monti S, Spina V, et al. Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica. 2016;101(4):e135–8.
Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–84.
Landau DA, Sun C, Rosebrock D, Herman SEM, Fein J, Sivina M, et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat Commun. 2017;8(1):2185.
• Ahn IE, Underbayev C, Albitar A, Herman SE, Tian X, Maric I, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017;129(11):1469–79 This study provided strong evidence for the role of subclonal heterogeneity and timing the rise of resistant disease in CLLs treated with ibrutinib.
Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45(3):290–4.
Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19(3):368–71.
Dieck CL, Tzoneva G, Forouhar F, Carpenter Z, Ambesi-Impiombato A, Sanchez-Martin M, et al. Structure and mechanisms of NT5C2 mutations driving thiopurine resistance in relapsed lymphoblastic leukemia. Cancer Cell. 2018;34(1):136–47 e6.
•• Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sanchez-Martin M, Madubata CJ, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553(7689):511–4 Systematic demonstration of clonal evolution in relapsed ALL and of the rise of resistant mutations at the cost of impaired leukemia cell growth.
Aries IM, van den Dungen RE, Koudijs MJ, Cuppen E, Voest E, Molenaar JJ, et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica. 2015;100(4):e132–6.
Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124(23):3420–30.
Irving JA, Enshaei A, Parker CA, Sutton R, Kuiper RP, Erhorn A, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2016;128(7):911–22.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2016;113(40):11306–11.
Schuurhuis GJ, Heuser M, Freeman S, Bene MC, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD working party. Blood. 2018;131(12):1275–91.
Thompson PA, Wierda WG. Eliminating minimal residual disease as a therapeutic end point: working toward cure for patients with CLL. Blood. 2016;127(3):279–86.
Ojamies PN, Kontro M, Edgren H, Ellonen P, Lagstrom S, Almusa H, et al. Monitoring therapy responses at the leukemic subclone level by ultra-deep amplicon resequencing in acute myeloid leukemia. Leukemia. 2017;31(5):1048–58.
• Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378(13):1189–99 Persistence of mutations during complete remission was associated with a significant rate of relapse, and it showed lack of correlation for CHIP-associated mutations with AML prognosis.
Morita K, Kantarjian HM, Wang F, Yan Y, Bueso-Ramos C, Sasaki K, et al. Clearance of somatic mutations at remission and the risk of relapse in acute myeloid leukemia. J Clin Oncol. 2018;36(18):1788–97.
Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269–85.
Altrock PM, Liu LL, Michor F. The mathematics of cancer: integrating quantitative models. Nat Rev Cancer. 2015;15(12):730–45.
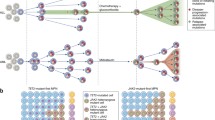
•• Beerenwinkel N, Schwarz RF, Gerstung M, Markowetz F. Cancer evolution: mathematical models and computational inference. Syst Biol. 2015;64(1):e1–25 A comprehensive review on methods to infer population dynamics and phylogenetic relationships in cancer.
Clapp G, Levy D. A review of mathematical models for leukemia and lymphoma. Drug Discov Today Dis Models. 2015;16:1–6.
Michor F, Beal K. Improving cancer treatment via mathematical modeling: surmounting the challenges is worth the effort. Cell. 2015;163(5):1059–63.
Rodriguez-Brenes IA, Wodarz D. Preventing clonal evolutionary processes in cancer: insights from mathematical models. Proc Natl Acad Sci U S A. 2015;112(29):8843–50.
Zhao B, Hemann MT, Lauffenburger DA. Modeling tumor clonal evolution for drug combinations design. Trends Cancer. 2016;2(3):144–58.
Komarova NL, Burger JA, Wodarz D. Evolution of ibrutinib resistance in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A. 2014;111(38):13906–11.
Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115(3):755–64.
Wodarz D, Garg N, Komarova NL, Benjamini O, Keating MJ, Wierda WG, et al. Kinetics of CLL cells in tissues and blood during therapy with the BTK inhibitor ibrutinib. Blood. 2014;123(26):4132–5.
Burger JA, Li KW, Keating MJ, Sivina M, Amer AM, Garg N, et al. Leukemia cell proliferation and death in chronic lymphocytic leukemia patients on therapy with the BTK inhibitor ibrutinib. JCI Insight. 2017;2(2):e89904.
Chen LS, Bose P, Cruz ND, Jiang Y, Wu Q, Thompson PA, et al. A pilot study of lower doses of ibrutinib in patients with chronic lymphocytic leukemia. Blood. 2018.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–28.
Caravagna G, Giarratano Y, Ramazzotti D, Tomlinson I, Graham TA, Sanguinetti G, et al. Detecting repeated cancer evolution from multi-region tumor sequencing data. Nat Methods. 2018;15(9):707–14.
Somarelli JA, Ware KE, Kostadinov R, Robinson JM, Amri H, Abu-Asab M, et al. PhyloOncology: understanding cancer through phylogenetic analysis. Biochim Biophys Acta Rev Cancer. 2017;1867(2):101–8.
Billera LJ, Holmes SP, Vogtmann K. Geometry of the space of phylogenetic trees. Adv Appl Math. 2001;27(4):733–67.
Zairis S, Khiabanian H, Blumberg A, Rabadan R. Moduli spaces of phylogenetic trees describing tumor evolutionary patterns. Lect Notes Comput Sci. 2014;8609:528–39.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–10.
Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22(7):792–9.
Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120(20):4191–6.
Zairis S, Khiabanian, H., Blumberg A, Rabadan R. Genomic data analysis in tree spaces. arXiv:1607.075032016.
Andrew J. Blumberg, Prithwish Bhaumik, Walker SG. Testing to distinguish measures on metric spaces. arXiv:1802.011522018.
Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–34.
Rosenbloom DIS, Camara PG, Chu T, Rabadan R. Evolutionary scalpels for dissecting tumor ecosystems. Biochim Biophys Acta Rev Cancer. 2017;1867(2):69–83.
Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–8.
Emmett KJ, Lee A, Khiabanian H, Rabadan R. High-resolution genomic surveillance of 2014 Ebolavirus using shared subclonal variants. PLoS Curr 2015;7.
Sobel Leonard A, Weissman DB, Greenbaum B, Ghedin E, Koelle K. Transmission bottleneck size estimation from pathogen deep-sequencing data, with an application to human influenza A virus. J Virol. 2017;91(14).
• Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747 A mathematical model showing that simultaneous therapy with two drugs is more effective than sequential therapy.
Acknowledgments
J-WL is a pre-doctoral fellow of the New Jersey Commission on Cancer Research (DFS18PPC017). HK acknowledges support from Rutgers Cancer Institute of New Jersey (P30CA072720).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Mathematical Models of Stem Cell Behavior
Rights and permissions
About this article
Cite this article
Loh, J.W., Khiabanian, H. Leukemia’s Clonal Evolution in Development, Progression, and Relapse. Curr Stem Cell Rep 5, 73–81 (2019). https://doi.org/10.1007/s40778-019-00157-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-019-00157-y