Abstract
Accurate and timely diagnosis is the first step for initiating effective treatment for tuberculosis (TB) and interrupting transmission. Worldwide, nearly one third of all TB cases go undetected or unreported each year. The emergence of extensively drug-resistant TB, in addition to challenges in detecting TB among children and people living with HIV has created an urgent need for better technologies. In the past 5 years, Xpert MTB/RIF has proved to be pathfinder for improved diagnosis. However, gaps remain. Currently, there is no molecular replacement for sputum smear microscopy at the level of peripheral laboratories. There is no simple, non-sputum-based biomarker test that can detect all forms of TB at the primary care level. There is also a need to align emerging TB drug regimens with appropriate companion diagnostics. This review describes advances in non-molecular and molecular diagnostics and their potential to fill the gaps in TB case detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB) is responsible for approximately 1.5 million deaths annually [1]. Worldwide, the number of multidrug-resistant TB (MDR-TB) cases has risen from 23,454 cases in 2006 to 480,000 in 2013 [1, 2•]. The post-2015 Global TB Strategy (“End TB Strategy”), proposed by the WHO and partners, aims to reduce incident cases of TB by 50 % and deaths by 75 % between 2015 and 2025, and reduce the burden of treatment and diagnosis costs due to TB [2•]. Ensuring accurate and timely case detection is critical to achieving these milestones, particularly as resistant strains become more prevalent [2•]. Ultimately, the End TB Strategy calls for universal access to drug-susceptibility testing for all patients at the time of TB detection.
There are three main issues that impede effective TB diagnosis. First, many patients in high-TB burden countries (HBCs) do not have access to effective diagnosis. The WHO estimates that in 2013 only 5.6 million of 9 million incident global cases (64 %) were detected and notified [1]. Second, despite the availability of modern diagnostics, many of these tools have limited use in HBCs, which are mostly low and middle income countries. The costs and infrastructural requirements of many technologies can be prohibitive. Third, there are limitations associated with existing diagnostics’ capacity to detect extrapulmonary tuberculosis (EPTB), pediatric TB, TB in people living with HIV (PLHIV), and drug resistance. Drug-susceptibility testing (DST) is necessary for the detection of drug resistance, and initiation of appropriate treatment for MDR-TB. However, in 2013, only 8.3 % of new cases and 17 % of re-treatment cases underwent DST [2•].
There are three main goals for advances in TB diagnostics: reduce diagnostic and treatment delays, enable point of care (POC) testing for same-day treatment, and support greater access to drug-susceptibility testing. Additional goals include reduced complexity, improving robustness, and improving accuracy.
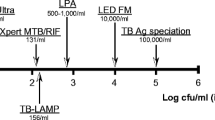
The 2014 edition of TB Diagnostics Technology and Market Landscape Report by UNITAID identified 191 TB diagnostic products that are either commercially available or in development. These technologies range from computer-aided diagnostics (CAD) used in conjunction with digital radiography to new biomarker-based techniques, such as volatile organic compounds (VOCs). Figure 1 shows the current TB diagnostics pipeline [3]. This review examines existing and novel, molecular and non-molecular diagnostic tools. It also explores their potential to increase access and availability, rapid case detection, and identify resistance.
Non-molecular Technologies
Novel Microscopy and Imaging Tools
Sputum smear microscopy (SSM), based on direct visualization of acid-fast bacilli, is the primary diagnostic method for identifying pulmonary TB in many HBCs [4]. Conventional SSM is low cost, has limited infrastructural needs, and trained microscopists can read 20–30 slides per day. However, microscopy has highly variable sensitivity (20–80 %) and poor accuracy among persons with HIV infection [5]. The use of fluorescent dyes over conventional staining enables easier reading at lower magnification, and fluorescent microscopy (FM) is about 10 % more sensitive than light microscopy [6]. Traditional FM microscopes require dark rooms and have high costs. These challenges have been addressed by light emitting diode (LED) microscopes, examples of which include the Primo Star iLED and CyScope® TB Fluorescence Microscope. Conventional microscopes can also be retrofitted with LED light sources for FM use.
The WHO has endorsed LED-FM for SSM over conventional light microscopy, and in HBCs, it has been found that there is a preference for use of LED-FM over traditional FM systems [7, 8]. Automated staining tools are used to reduce workloads and improve staining results. For example, the RAL STAINER (Ral Diagnostics, Martillac, France) and Aerospray TB Series 2 (ELITechGroup, Estavayer-le-Lac, Switzerland) rapidly process slides using fluorescent dyes.
Recent innovations have focused on integrated systems in which stained slides are automatically loaded into a microscope, digital imagery is acquired, and images are analyzed using propriety algorithms. One example, the TBDx system (Applied Visual Sciences, Inc, Leesburg, USA) has shown performance comparable to manual examination of FM slides by trained microscopists for positive and negative, but not scanty, slides [9]. The use of digital imagery, mobile, and tablet technology, which are increasingly outfitted with high-resolution cameras, has opened new potential for low-cost microscopy. For example, a tablet camera attached to an optical chamber with LED filters has been used to read slides. The device has shown sensitivities within 15 % of LED-FM devices [10]. In addition to facilitated use and portability, the use of digital and mobile technology allows for image archival and automated reading algorithms.
Chest x-rays have high sensitivity for TB (80–95 %), but modest specificity (70–75 %), as several other lung diseases can cause x-ray abnormalities. Therefore, chest x-ray is a good triage or screening tool, and can identify those who require confirmatory testing. The use of automated reading algorithms has become important in the field of chest radiography. Radiographic tools are being revolutionized in two ways: first, through the use of digital plates and digital radiography, and second through computer-aided diagnosis [11]. Direct digital radiography consists of a digital detector which replaces the plates present in conventional radiography; computer radiography involves a specific plate which produces an analog signal allowing the image to be digitized and stored on a dedicated computer [12]. Although the upfront costs of conventional radiography are less, the use of digital radiography is more cost-efficient over the long term by removing recurring costs, eliminating reagent use, and decreasing radiation dose. Further, digitized radiography allows for easier and inexpensive archiving and transmission of data, although it must be noted that robust data transmission capacity is necessary.
The reproducibility of radiographic results is an issue that Computer-aided Diagnosis (CAD) software has sought to address by providing objective interpretation of these results [13]. An excellent example of the CAD software is the recently developed CAD4TB by Diagnostic Image Analysis Group, Nijmegen, Netherlands. This software can be applied to digital radiography and to conventional chest X-ray films which are electronically scanned and analyzed. CAD and human readers have comparable inter-reader agreement levels [13]. A study from Zambia reported that the use of CAD with digital radiographs in areas where radiologists are not available to interpret chest x-rays has the potential to make chest radiography screening accessible [2•].
Culture-Based Technologies
Cultures are the gold standard for TB diagnosis and DST. Cultures are sensitive and can detect low levels of Mycobacterium tuberculosis (MTB) [14]. However, they have important limitations. First, culture-based diagnosis for TB has a slow turnaround time. Second, trained personnel are required for the processing and monitoring of cultures. Third, culture specimens require biosafety level 3 environments.
Solid cultures are time-consuming and provide limited clinical value. Conversely, the use of liquid culture media allows for more rapid results, obtained in 10–14 days, in addition to up to 10 % more sensitivity compared to solid cultures [15]. Liquid cultures therefore play an important role in identifying microbial resistance to rifampicin (RIF), isoniazid (INH), pyrazinamide (PZA), ethambutol (EMB), and streptomycin (STR). Automated liquid culture platforms, specifically the mycobacterial growth indicator tube Bactec MGIT (BD Diagnostics, New Jersey, USA), which has been endorsed by the WHO for DST, scans processed MGIT sputum bottles and determines MTB growth by measuring O2 depletion [16]. Another system, BacT/Alert 3D (bioMérieux, Marcy l’Etoile, France) measures MTB levels based on CO2 accumulation. Liquid culture systems are, however, more complex and expensive than solid culture and can have high contamination rates.
Microscopically observed drug susceptibility (MODS), a manual culture assay, has been shown to perform as well as automated liquid culture systems, and can be used as a rapid test to identify MTB and resistance to RIF and INH [17]. Other manual non-commercial tests include colorimetric redox indicator (CRI), nitrate reductase assay (NRA), and the thin layer agar (TLA) assay. The limitations of these non-commercial tests are that users require extensive training in order to ensure standardization and quality, rendering scalability impractical.
Biomarker-Based Technologies
Non-sputum-based tests are particularly important for diagnosing pediatric TB patients, extrapulmonary TB patients, and HIV comorbid patients. Pediatric TB accounts for approximately 11 % of TB cases globally, and in HBCs account for nearly 80 % of incidents of childhood cases [18]. Pediatric TB is challenging to diagnose as it is very difficult to obtain sufficient sputum from children. Even in cases where sufficient sputum is obtained, culture-based methods have low yields (30–40 %) [19]. HIV comorbidity with TB also represents a diagnostic challenge. The presence of HIV not only worsens TB outcomes, but makes TB more difficult to diagnose using conventional sputum-based diagnostics. Also, higher rates of extrapulmonary TB are observed in those co-infected with HIV [20].
Biomarker-based technologies may be ideal for diagnosing TB in these difficult to diagnose populations. While existing serological, antibody tests have been shown to lack accuracy and are discouraged by the WHO [21], research in this area is important, as serological tests might open the door for a simple, rapid, lateral flow test. MBio Diagnostics Inc (Boulder, USA), and TB Biosciences, Bethlehem, USA, are among several groups developing novel antibody-based technologies.
Antigen detection might also hold some promise (e.g., lipoarabinomannan (LAM) in urine). The Alere Determine TB LAM Ag rapid test (Alere Inc., Waltham, USA) allows for detection of LAM and aids in the detection of TB in HIV-infected patients who are severely immune-suppressed. The LAM assay is a simple strip test that is easy to perform. However, LAM assays have low sensitivity in patients with CD4 counts greater than 200 cells/\( \upmu \)l [22], and their use is not endorsed by the WHO. A challenge associated with the LAM assay is the fact that it is not specific for MTBC. Enzymatic detection methods are also under development. Global BioDiagnostics Corp, Texas, USA, has developed a biophotonic detection platform that utilizes reporter enzyme fluorescence to detect β-lactamase-producing live TB bacteria in sputum samples.
Volatile organic compounds (VOCs) allow for the detection of early MTB infections through breath. The use of VOCs might allow for improved diagnosis of MTB in children and PLHIV. The feasibility of using VOCs as a diagnostic tool has been shown using giant African pouched cane rats, trained to identify VOCs from TB-infected sputum. In this proof-of-concept study, VOCs were shown to have better performance when compared to smear microscopy [8, 23]. Commercial VOC-based technologies, for example Aenose (eNose Company, Zutphen, The Netherlands), BreathLink (Menssana Research Inc, Newark, USA), and TB breathalyzer (Rapid Biosensor Systems, Cambridge, UK) are under development or evaluation. However, there are limited data to support their clinical use at this time.
Extrapulmonary tuberculosis (EPTB) can affect almost any site of the body, including body fluids (plural, pericardial, and ascitic fluids), CNS, and lymph nodes. Conventional culture methods perform poorly for EPTB, for example MTB detection in pleural TB is 24–58 % [24]. However, biomarkers including adenosine deaminase (ADA) and free interferon gamma (IFN-g) have the potential to detect EPTB, especially pleural TB [25].
The diagnosis and treatment of latent TB infection is important in specific high-risk populations such as people living with HIV, contacts of TB cases, persons initiating anti-tumor necrosis factor (TNF-alpha) treatment, patients with end-stage renal failure on dialysis, and patients preparing for organ or hematologic transplantation. There are two main tests used to screen for latent tuberculosis infection (LTBI): tuberculin skin test (TST) and interferon gamma release assay (IGRA).
Many countries have two IGRAs available, QuantiFERON-TB Gold in Tube (Qiagen, Valencia, USA) and T-SPOT TB assay (Oxford Immunotec, Abingdon, UK) [26]. Both TST and IGRA are acceptable; however, they each present challenges. Both have reduced sensitivity in immune-suppressed persons, and cannot separate latent from active TB [26]. Both tests have only modest predictive value to detect those who are at highest risk of progression from latent to active TB [27]. Available longitudinal data suggest that a majority (>95 %) of those with positive TST or IGRA results do not progress to active TB disease during follow-up [27].
Newer LTBI tests are being developed to overcome the limitations of existing tests. In 2015, Qiagen announced QuantiFERON®-TB Gold Plus (QFT®-Plus), the fourth-generation IGRA which incorporates CD8+ T cell response data. Autoimmune Diagnostika (Straßberg, Germany) [2•] is currently developing an IGRA and IL-2 assay, which can distinguish between active and latent TB. ProteinLogic is developing ImmiPrint® (ProteinLogic, Cambridge, UK) [2•], a multiplexed antibody assay which gives a finger print to active and latent infection antigens, facilitating the differentiation between the two forms of tuberculosis. None of these newer LTBI assays have been validated yet, nor approved for clinical use.
Molecular Techniques
Nucleic acid amplification tests (NAATs) amplify genome-specific targets, through different methods: polymerase chain reaction (PCR), transcription-mediated amplification (TMA), loop-mediated amplification (LAMP), nucleic acid sequence-based amplification (NASBA), and strand displacement amplification (SDA). Figure 2 shows the TB NAAT pipeline, including tools that are under development [2•, 3].
Line Probe Assays
Line probe assays were the first TB NAATs to be endorsed by the WHO in 2008. They extract genomic DNA, then select DNA targets, and label them with biotin to create amplicons. The amplicons are then applied to strips with specific oligonucleotide probes. Subsequently, bound amplicons are detected through attachment of a label and the visualization of a dark band against a scoring chart. These assays allow for identification of mutations, which aids in drug-susceptibility testing. Based on a meta-analysis, GenoType MTBDRplus line probe assays (LPA) (Hain LifeScience, Nehren, Germany) has 98.1 % sensitivity and 98.7 % specificity for RIF resistance, and 84.3 % sensitivity and 99.5 % specificity for INH resistance [28]. Due to the high sensitivity of the test, any contamination may alter test results, thus limiting the use of LPAs in reference or intermediate laboratories [2•]. In addition, highly trained personnel are required to run the assay.
Currently, the Genotype MTBDRplus assay is endorsed by the WHO for rapid detection of MDR-TB in smear-positive samples and culture isolates, and special negotiated prices are available for HBCs. Another test, the Genotype MTB-DRsl, allows for the detection of resistance of second-line drugs to increase the diagnosis of XDR-TB and MDR-TB. Based on a meta-analysis, this LPA has 76.9 % sensitivity and 99.5 % specificity for indirect injection methods, and 98.0 % sensitivity and 97.8 % specificity for direct injection methods [29]. This LPA for second-line drugs has not been endorsed by the WHO.
Commercial NAATs for Use in Reference and Intermediate-tier Laboratories
Originally the first NAATs, designed for reference and intermediate-tier laboratories, targeted high-income countries and their markets. These include Hologic Gen-Probe® (San Diego, USA) and COBAS® TaqMan® MTB test (Roche Molecular Diagnostics, Pleasanton, USA) [1, 30]. There are other commercial tests employed in reference laboratories; however, none have been endorsed by the WHO.
Microarray Platforms
Several microarray platforms are under development, these include VereMTB detection kit (Veredus Laboratories, Singapore), iCubate (Huntsville, USA), and Hydra 1K (Stanford University, Palo Alto, USA, and Insilixa, Sunnyvale, USA) [2•]. Akonni Biosystems, Frederick, USA, has developed TruArray which is used for MDR-TB diagnosis. It is able to distinguish between MTB and Mycobacterium avium through the use of a single tube PCR reaction, which incorporates a fluorescent label to facilitate visualization. CapitalBio Corp (Beijing, China) also produces microarray products for detecting RIF resistance in culture isolates and sputum samples [31]. None of these microarray products are adequately validated and none are endorsed by the WHO.
Modular, Cartridge-Based, Automated NAATs
Automated, cartridge-based NAATs simplify and automate all the steps involved in NAATs, and enhance biosafety and ease of use. The first such TB NAAT is the Xpert MTB/RIF (Cepheid Inc, USA). FIND along with the University of Medicine and Dentistry of New Jersey collaborated with Cepheid to create Xpert, a combined TB and RIF resistance assay. All parts of the test are performed on one cartridge, which is able to produce results in approximately 2 h [32•]. The WHO endorsed Xpert in 2013 indicating that it should be used instead of conventional microscopy and culture for initial diagnosis in adults and children suspected with TB. Since its introduction, more than 10 million Xpert cartridges have been used in HBCs, where the cartridges are available at a special price of $9.98. Based on a meta-analysis, Xpert has a sensitivity of 89 % and a specificity of 99 % for detecting pulmonary TB, and a sensitivity of 95 % and specificity of 98 % for RIF resistance [33•]. The WHO has also endorsed Xpert for specific types of extrapulmonary samples (i.e., CSF, lymph nodes, and tissues) based on good accuracy data [34]. Recently, Cepheid Inc. announced the development of Xpert® MTB/RIF Ultra, a next-generation test with increased sensitivity to aid TB detection in smear-negative patients.
NAATs Developed for Use in Lower-tier Laboratories and Microscopy Centers
Although Xpert is currently being rolled out in multiple countries, most high-TB burden countries are primarily using it as a rapid DST tool, mainly due to high cost [35]. The technology is challenging to deploy at the microscopy center level. Several companies are developing tools that can be used in lower-tier laboratories, specifically in high-TB burden countries. Such tools include the TrueNAT assay (Mobio Inc, Goa, India), Genedrive platform (Epistem, Manchester, UK), and EasyNAT (Ustar Biotechnologies, Hangzhou, China). While these products are already on the market, the evidence base is weak, and further evaluations are necessary for policy development.
Building on its POC CD4 and viral load assays, Alere Inc, Waltham, USA is developing the Alere q assay for rapid and accurate diagnosis of TB while performing drug resistance testing as well. This assay will have the ability to detect resistance to multiple drugs. GenePOC Inc (Canada) is creating a real-time PCR which can detect MTBC and resistance for rifampicin (RIF) and fluoroquinolone (FLQ). GenePOC Inc is innovating this platform to visualize its results on a touch screen interface [2•]. QuantuMDx is developing MTBC and MDR assays using a handheld unit which will use a graphical interface to view results. This assay permits the ability to simultaneously test a large number of subjects using a small and inexpensive cartridge.
Sequencing Methods
Next-generation sequencing (NGS) methods have allowed for large amounts of DNA data to be processed, profiled, characterized, and evaluated for protein interactions. These technologies are enabling both high-throughput and low-cost DNA sequencing, and will be particularly useful for assessing resistance to newer TB drugs. Shotgun sequencing is being used with next-generation methods. For example, research groups have used shotgun metagenomic techniques to detect and characterize MTBC on the platform MiSeq (Illumina, USA), a next-generation sequencing platform [36]. NGS also have a lot of potential for diagnostic and broader surveillance. A molecular epidemiologic study found that using sequencing information gave better contact tracing information regarding TB infection compared to classic genotyping [37].
Whole genome sequencing is allowing for the identification of de novo mutations associated with phenotypic drug resistance. For example, a study found that second-line drugs were exerting selection effects for mutations which induce drug resistance [38]. Characterizing these new resistant strains will allow for the design of diagnostics that can screen for specific mutations. Novel sequencing techniques also allow for the detection of heteroresistance, which may be misclassified using non-molecular methods. Accompanying these tools are computational algorithms to analyze data, and cloud-based systems to share data with larger data repositories.
Market Landscape and Unmet Needs
In the past 5 years, the successful roll out of Xpert MTB/RIF has generated great interest in the development of new TB diagnostics. The current diagnostics market is one thriving based on strong public and private partnerships. For example, by December 2014, over 10 million cartridges of Xpert were made available to the public sector in more than 100 countries [39]. The highest number of tests has so far been procured by South Africa. The roll out of Xpert has also been quite successful in China, India, and Brazil. Despite these successes, gaps remain in the area of TB diagnostics. Xpert is expensive, not designed for lower tiers of the healthcare system, and can only detect resistance against rifampicin [35].
It is therefore critical to promote the introduction of new products, and identify areas where the incremental value of new technologies and the potential market is the greatest. Kik and colleagues developed a series of target product profiles and how they scored with respect to prioritization by stakeholders, impact, market, implementation, and scalability [40•]. Based on this needs assessment exercise, the following diagnostics have been identified as the highest priorities for future research and development:
-
A rapid, sputum-based test that can replace sputum smear microscopy
-
A point of care, non-sputum-based test using biomarkers common to all forms of TB
-
A rapid triage test, for use by first-contact health care providers, to identify people suspected of having TB
-
A rapid drug-susceptibility test at the level of microscopy centers
For all of the above priorities, detailed target product profiles have been developed, and reviewed by a WHO expert consensus group [41•]. In addition, a series of market analyses studies have been published to provide product developers the information they need to make investments in new tool development [4, 42, 43]. A companion website has been created to provide information useful to TB product developers (www.tbfaqs.org). The engagement of product developers, TB care providers, and other stakeholders ensures a steady product pipeline, and appropriate assessment of these products demonstrates that the field of TB diagnostics is ripe with opportunities to develop new tools that overcome current diagnostic challenges.
Conclusion
Gaps in TB case finding and the emergence of drug-resistant TB have created a pressing need for robust and accurate diagnostics. Newer tests such as Xpert MTB/RIF are having an impact in increasing case detection and reducing time to treatment. Their use should steadily increase and enable countries to reach the goal of universal DST. After several decades, new TB drugs have been introduced. These include bedaquiline (Sirturo® by Janssen Therapeutics), and delamanid (Deltyba® by Otsuka Pharmaceutical Co., Ltd.). Phase 3 trials are underway to evaluate a new drug regimen called PaMZ (which contains Pretomanid® (previously called PA-824), moxifloxacin, and pyrazinamide). If successful, the TB community will gear up to introduce new, shorter treatment regimens in the next 5 years. National tuberculosis programs will need to streamline regulatory and policy adoption processes, and prepare the ground for the launch of these new regimens. It is critical to make sure that new regimens are introduced along with companion diagnostics that can assess drug resistance to new drugs and regimens. A comprehensive “test and treat” strategy is critical for making progress towards the End TB goal.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
World Health Organization, Global Tuberculosis Report 2014. 2014: World Health Organization.
UNITAID, Tuberculosis: diagnostic, technology and market landscape. 2014, WHO: Geneva. p. 1–42. This report identifies and reviews over 191 technologies at various stages in the TB diagnostics pipeline, and characterizes potential opportunities for future market entry and innovation.
Pai M, Schito M. Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis. 2015;211(S2):S21–8.
Kik SV, Denkinger CM, Chedore P, Pai M. Replacing smear microscopy for the diagnosis of tuberculosis: what is the market potential? Eur Respir J. 2014;43(6):1793–6.
Mugusi F, Villamor E, Urassa W, Saathoff E, Bosch R, Fawzi W. HIV co-infection, CD4 cell counts and clinical correlates of bacillary density in pulmonary tuberculosis. Int J Tuberc Lung Dis. 2006;10(6):663–9.
Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, Cunningham J, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(9):570–81.
Albert H, Nakiyingi L, Sempa J, Mbabazi O, Mukkada S, Nyesiga B, et al. Operational implementation of LED fluorescence microscopy in screening tuberculosis suspects in an urban HIV clinic in Uganda. PLoS One. 2013;8(9):e72556.
Mgode GF, Weetjens BJ, Nawrath T, Cox C, Jubitana M, Machang’u RS, et al. Diagnosis of tuberculosis by trained African giant pouched rats and confounding impact of pathogens and microflora of the respiratory tract. J Clin Microbiol. 2012;50(2):274–80.
Lewis JJ, Chihota VN, van der Meulen M, Fourie PB, Fielding KL, Grant AD, et al. “Proof-Of-Concept” evaluation of an automated sputum smear microscopy system for tuberculosis diagnosis. PLoS One. 2012;7(11):e50173.
Tapley A, Switz N, Reber C, Davis JL, Miller C, Matovu JB, et al. Mobile digital fluorescence microscopy for diagnosis of tuberculosis. J Clin Microbiol. 2013;51(6):1774–8.
World Health Organization, Tuberculosis prevalence surveys: a handbook. 2011.
Cruz R. Digital radiography, image archiving and image display: practical tips. Can Vet J. 2008;49(11):1122.
Muyoyeta M, Maduskar P, Moyo M, Kasese N, Milimo D, Spooner R, et al. The sensitivity and specificity of using a computer aided diagnosis program for automatically scoring chest X-rays of presumptive TB patients compared with Xpert MTB/RIF in Lusaka Zambia. PLoS One. 2014;9(4):e93757.
Jr Yeager H, Lacy J, Smith L, LeMaistre C. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95(6):998.
Cruciani M, Scarparo C, Malena M, Bosco O, Serpelloni G, Mengoli C. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J Clin Microbiol. 2004;42(5):2321–5.
World Health Organization. Use of liquid TB culture and drug susceptibility testing (DST) in low and medium income settings. Geneva: World Health Organization; 2007.
Minion J, Leung E, Menzies D, Pai M. Microscopic-observation drug susceptibility and thin layer agar assays for the detection of drug resistant tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10(10):688–98.
World Health Organization, Guidance for national tuberculosis programmes on the management of tuberculosis in children. 2014: World Health Organization.
Marais BJ, Pai M. New approaches and emerging technologies in the diagnosis of childhood tuberculosis. Paediatr Respir Rev. 2007;8(2):124–33.
El-Sadr WM, Tsiouris SJ. HIV-associated tuberculosis: diagnostic and treatment challenges. in Seminars in respiratory and critical care medicine. 2008.
Steingart KR, Flores LL, Dendukuri N, Schiller I, Laal S, Ramsay A, et al. Commercial serological tests for the diagnosis of active pulmonary and extrapulmonary tuberculosis: an updated systematic review and meta-analysis. PLoS Med. 2011;8(8):e1001062.
Minion J, Leung E, Talbot E, Dheda K, Pai M, Menzies D. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur Respir J. 2011;38(6):1398–405.
Phillips M, Basa-Dalay V, Bothamley G, Cataneo RN, Lam PK, Natividad MPR, et al. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis. 2010;90(2):145–51.
Trajman A, Pai M, Dheda K, van Zyl Smit R, Zwerling A, Joshi R, et al. Novel tests for diagnosing tuberculous pleural effusion: what works and what does not? Eur Respir J. 2008;31(5):1098–106.
Greco S, Girardi E, Masciangelo R, Capoccetta G, Saltini C. Adenosine deaminase and interferon gamma measurements for the diagnosis of tuberculous pleurisy: a meta-analysis. Int J Tuberc Lung Dis. 2003;7(8):777–86.
Pai M, Denkinger CM, Kik SV, Rangaka MX, Zwerling A, Oxlade O, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev. 2014;27(1):3–20.
Rangaka MX, Wilkinson KA, Glynn JR, Ling D, Menzies D, Mwansa-Kambafwile J, et al. Predictive value of interferon-γ release assays for incident active tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):45–55.
Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 2008;32(5):1165–74.
Theron GJ, Peter M, Richardson M, Barnard S, Donegan R, Warren KR, et al. The diagnostic accuracy of the GenoType® MTBDRsl assay for the detection of resistance to second‐line anti‐tuberculosis drugs. The Cochrane Library. 2014.
World Health Organization, Global tuberculosis control: WHO report: World Health Organization Global Tuberculosis Programme. 2008.
Weyer K, Mirzayev F, Migliori GB, van Gemert W, D’Ambrosio L, Zignol M, et al. Rapid molecular TB diagnosis: evidence, policy making and global implementation of Xpert MTB/RIF. Eur Respir J. 2013;42(1):252–71.
World Health Organization, Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MT. 2013.
Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. The Cochrane Library. 2014.
Denkinger CM, Schumacher SG, Boehme CC, Dendukuri N, Pai M, Steingart KR. Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. European Respiratory Journal. 2014: p. erj00078-2014.
Qin ZZ, Pai M, Van Gemert W, Sahu S, Ghiasi M, Creswell J. How is Xpert MTB/RIF being implemented in 22 high tuberculosis burden countries? European Respiratory Journal. 2014: p. erj01477-2014.
Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ. 2014. 2: p. e585.
Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10(2):e1001387.
Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013;8(12):e82551.
World Health Organization. TB diagnostics and laboratory strengthening. 2015 [cited 2015 10 Feb]; Available from: http://www.who.int/tb/laboratory/mtbrifrollout/en/.
Kik SV, Denkinger CM, Casenghi M, Vadnais C, Pai M. Tuberculosis diagnostics: which target product profiles should be prioritised? Eur Respir J. 2014;44(2):537–40. This article constructs a useful prioritization scheme for target product profiles of new diagnostics based on feedback from a wide range of stakeholder groups, and examines areas of consensus regarding development priorities.
World Health Organization, High priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting, 28–29 April 2014, Geneva, Switzerland. 2014. This report details outcomes of a consensus-gathering meeting in Geneva (April 2014) to identify the critical characteristics of target product profiles and set priorities for future diagnostics.
TB Diagnostics Market Analysis Consortium, Market assessment of tuberculosis diagnostics in Brazil in 2012. PloS one. 2014. 9;8: p. e104105.
Pai M, Palamountain K. New tuberculosis technologies: challenges for retooling and scale-up [State of the art series. New tools. Number 4 in the series]. Int J Tuberc Lung Dis. 2012;16(10):1281–90.
Compliance with Ethics Guidelines
Conflict of Interest
Marzieh Ghiasi, Tripti Pande, and Madhukar Pai declare that they have no conflict of interest. M. Pai serves as a consultant to the Bill and Melinda Gates Foundation and serves on the Scientific Advisory Committee of FIND, Geneva.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Bacterial Tropical Medicine
Marzieh Ghiasi and Tripti Pande contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ghiasi, M., Pande, T. & Pai, M. Advances in Tuberculosis Diagnostics. Curr Trop Med Rep 2, 54–61 (2015). https://doi.org/10.1007/s40475-015-0043-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40475-015-0043-1