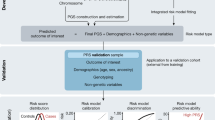
Abstract
Purpose of Review
To explain how the intended use of polygenic risk scores (PRSs) in healthcare guides the design and evaluation of prediction studies.
Recent Findings
The advances in gene discovery in common complex diseases have fueled the interest in the potential of PRSs to predict risks and improve the prevention and early detection of disease. As the predictive ability of a PRS differs between populations and settings, it is important that prediction studies are designed and evaluated with the intended use of the risk scores in mind, but this is rarely done.
Summary
The intended use indicates in whom and how the PRS will be used in healthcare and for what purpose. This intended use dictates what outcome needs to be predicted in which population using which predictors. It also tells which other variables or clinical risk models might be available to improve the prediction. The intended use also provides the necessary context to evaluate whether the predictive ability of the PRS or the risk model that includes PRS is high enough for the score to be potentially useful in healthcare. The intended use should be leading risk prediction research.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
• Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24. https://doi.org/10.1038/s41588-018-0183-z This paper revived the interest in polygenic prediction as it concluded that PRSs can identify indiviuals at increased risk comparable to that of monogenic disorders. The methodology of the paper has been challenged in several commentaries, including [32, 33, and 34].
Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, et al. Genomic risk prediction of coronary artery disease in 480,000 adults. J Am Coll Cardiol. 2018;72:1883–93.
Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–13. https://doi.org/10.1038/s41588-018-0241-6.
Brautbar A, Pompeii LA, Dehghan A, Ngwa JS, Nambi V, Virani SS, et al. A genetic risk score based on direct associations with coronary heart disease improves coronary heart disease risk prediction in the Atherosclerosis Risk in Communities (ARIC), but not in the Rotterdam and Framingham Offspring, studies. Atherosclerosis. 2012;223:421–6.
Havulinna AS, Kettunen J, Ukkola O, Osmond C, Eriksson JG, Kesäniemi YA, et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32,669 individuals. Hypertens (Dallas, Tex 1979). 2013;61:987–94.
Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400.
Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–32.
Park HY, Choi HJ, Hong Y-C. Utilizing genetic predisposition score in predicting risk of type 2 diabetes mellitus incidence: a community-based cohort study on middle-aged Koreans. J Korean Med Sci. 2015;30:1101–9.
Matejcic M, Saunders EJ, Dadaev T, et al. Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat Commun. 2018;9:4616.
Natarajan P, Peloso GM, Zekavat SM, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun. 2018;9:3391.
Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–90.
Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9:665–74.
Haddow J, Palomaki G (2004) ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury M, Little J, Burke W editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. 217–233.
Yang X, Leslie G, Gentry-Maharaj A, Ryan A, Intermaggio M, Lee A, et al. Evaluation of polygenic risk scores for ovarian cancer risk prediction in a prospective cohort study. J Med Genet. 2018;55:546–54.
Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109. https://doi.org/10.1093/jnci/djw302.
Lecarpentier J, Silvestri V, Kuchenbaecker KB, Barrowdale D, Dennis J, McGuffog L, et al. Prediction of breast and prostate Cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol. 2017;35:2240–50.
Evans DG, Brentnall A, Byers H, Harkness E, Stavrinos P, Howell A, et al. The impact of a panel of 18 SNPs on breast cancer risk in women attending a UK familial screening clinic: a case–control study. J Med Genet. 2017;54:111–3.
Cust AE, Drummond M, Kanetsky PA, Goldstein AM, Barrett JH, MacGregor S, et al. Assessing the incremental contribution of common genomic variants to melanoma risk prediction in two population-based studies. J Invest Dermatol. 2018;138:2617–24.
Moons KGM, Royston P, Vergouwe Y, Grobbee DE, Altman DG. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:–b375.
Kim S-H, Lee E-S, Yoo J, Kim Y. Predicting risk of type 2 diabetes mellitus in Korean adults aged 40-69 by integrating clinical and genetic factors. Prim Care Diabetes. 2018;13:3–10. https://doi.org/10.1016/j.pcd.2018.07.004.
Chouraki V, Reitz C, Maury F, et al. Evaluation of a genetic risk score to improve risk prediction for Alzheimer’s disease. J Alzheimers Dis. 2016;53:921–32.
Zhang X, Rice M, Tworoger SS, Rosner BA, Eliassen AH, Tamimi RM, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: a nested case-control study. PLoS Med. 2018;15:e1002644.
Szulkin R, Whitington T, Eklund M, Aly M, Eeles RA, Easton D, et al. Prediction of individual genetic risk to prostate cancer using a polygenic score. Prostate. 2015;75:1467–74.
Chan CHT, Munusamy P, Loke SY, Koh GL, Yang AZY, Law HY, et al. Evaluation of three polygenic risk score models for the prediction of breast cancer risk in Singapore Chinese. Oncotarget. 2018;9:12796–804.
Tang EYH, Harrison SL, Errington L, Gordon MF, Visser PJ, Novak G, et al. Current developments in dementia risk prediction modelling: an updated systematic review. PLoS One. 2015;10:e0136181.
Lee C. Best linear unbiased prediction of individual polygenic susceptibility to sporadic vascular dementia. J Alzheimers Dis. 2016;53:1115–9.
Einziger T, Levi L, Zilberman-Hayun Y, Auerbach JG, Atzaba-Poria N, Arbelle S, et al. Predicting ADHD symptoms in adolescence from early childhood temperament traits. J Abnorm Child Psychol. 2018;46:265–76.
Bussu G, Jones EJH, Charman T, Johnson MH, Buitelaar JK, Team BASIS. Prediction of autism at 3 years from behavioural and developmental measures in high-risk infants: a longitudinal cross-domain classifier analysis. J Autism Dev Disord. 2018;48:2418–33.
Smith T, Gunter MJ, Tzoulaki I, Muller DC. Brief communication: the added value of genetic information in colorectal cancer risk prediction models: development and evaluation in the UK Biobank prospective cohort study. Br J Cancer. 2018. https://doi.org/10.1038/s41416-018-0282-8.
Khawaja AP, Cooke Bailey JN, Wareham NJ, et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50:778–82.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779.
Janssens ACJW, Joyner MJ. Polygenic risk scores that predict common diseases using millions of single nucleotide polymorphisms: is more, better? Clin Chem. 2019;2018:296103.
Greenland P, Hassan S. Precision preventive medicine—ready for prime time? JAMA Intern Med. 2019. https://doi.org/10.1001/jamainternmed.2019.0142.
Curtis D (2019) Clinical relevance of genome-wide polygenic score may be less than claimed. Ann Hum Genet ahg.12302. https://doi.org/10.1111/ahg.12302.
Ganna A, Ingelsson E. 5 year mortality predictors in 498 103 UK Biobank participants: a prospective population-based study. Lancet. 2015;386:533–40.
Jacobsen LM, Larsson HE, Tamura RN, Vehik K, Clasen J, Sosenko J, et al. Predicting progression to type 1 diabetes from ages 3 to 6 in islet autoantibody positive TEDDY children. Pediatr Diabetes. 2019;20:263–70. https://doi.org/10.1111/pedi.12812.
Martinelli V, Dalla Costa G, Messina MJ, di Maggio G, Sangalli F, Moiola L, et al. Multiple biomarkers improve the prediction of multiple sclerosis in clinically isolated syndromes. Acta Neurol Scand. 2017;136:454–61.
Pitkänen N, Juonala M, Rönnemaa T, Sabin MA, Hutri-Kähönen N, Kähönen M, et al. Role of conventional childhood risk factors versus genetic risk in the development of type 2 diabetes and impaired fasting glucose in adulthood: the Cardiovascular Risk in Young Finns Study. Diabetes Care. 2016;39:1393–9.
Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–38.
Van Calster B, Vickers AJ. Calibration of risk prediction models. Med Decis Mak. 2015;35:162–9.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–35.
Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308:1552.
Altman DG, Bland JM. Diagnostic tests 2: predictive values. BMJ. 1994;309:102.
Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–43.
Pencina MJ, D’Agostino RB, D’Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–72.
•• Cook NR. Quantifying the added value of new biomarkers: how and how not. Diagnostic Progn Res. 2018;2:14 This commentary provides an overview of methods currently used to evaluate new biomarkers, describes their strengths and limitations, and offers suggestions on their use.
Mihaescu R, van Zitteren M, van Hoek M, Sijbrands EJG, Uitterlinden AG, Witteman JCM, et al. Improvement of risk prediction by genomic profiling: reclassification measures versus the area under the receiver operating characteristic curve. Am J Epidemiol. 2010;172:353–61.
Martens FK, Tonk ECM, Janssens ACJW. Evaluation of polygenic risk models using multiple performance measures: a critical assessment of discordant results. Genet Med. 2018;21:391–7. https://doi.org/10.1038/s41436-018-0058-9.
Martens FK, Kers JG, Janssens ACJW. External validation is only needed when prediction models are worth it (Letter commenting on: J Clin Epidemiol. 2015;68:25–34). J Clin Epidemiol. 2016. https://doi.org/10.1016/j.jclinepi.2015.01.022.
Moons KG, Kengne AP, Grobbee DE, Royston P, Vergouwe Y, Altman DG, et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–8.
Abraham G, Havulinna AS, Bhalala OG, Byars SG, de Livera AM, Yetukuri L, et al. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–78.
Vassos E, Di Forti M, Coleman J, et al. An examination of polygenic score risk prediction in individuals with first-episode psychosis. Biol Psychiatry. 2017;81:470–7.
Selzam S, Krapohl E, von Stumm S, O’Reilly PF, Rimfeld K, Kovas Y, et al. Predicting educational achievement from DNA. Mol Psychiatry. 2017;22:267–72.
Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–23.
Gu F, Chen T-H, Pfeiffer RM, Fargnoli MC, Calista D, Ghiorzo P, et al. Combining common genetic variants and non-genetic risk factors to predict risk of cutaneous melanoma. Hum Mol Genet. 2018;27:4145–56.
•• Wynants L, Collins GS, Van Calster B. Key steps and common pitfalls in developing and validating risk models. BJOG. 2017;124:423–32 This paper presents an overview of ten steps from the conception of the study to the implementation of the risk model and discusses common pitfalls.
Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63.
•• Janssens ACJW, Ioannidis JPA, Bedrosian S, Boffetta P, Dolan SM, Dowling N, et al. Strengthening the reporting of Genetic RIsk Prediction Studies (GRIPS): explanation and elaboration. J Clin Epidemiol. 2011;64:e1–e22 This paper provides recommendations to enhance the transparency, completeness, and accuracy of reporting genetic prediction studies.
Funding
This work was supported by a consolidator grant from the European Research Council (Genomic Medicine).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Epidemiologic Methods
Rights and permissions
About this article
Cite this article
Martens, F.K., Janssens, A.C.J. How the Intended Use of Polygenic Risk Scores Guides the Design and Evaluation of Prediction Studies. Curr Epidemiol Rep 6, 184–190 (2019). https://doi.org/10.1007/s40471-019-00203-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40471-019-00203-7