Abstract
Background and Objective
Pediatric cardiomyopathies are clinically heterogeneous heart muscle disorders associated with significant morbidity and mortality for which substantial evidence for a genetic contribution was previously reported. We present a detailed molecular investigation of a cohort of 231 patients presenting with primary cardiomyopathy below the age of 18 years.
Methods
Cases with pediatric cardiomyopathies were analyzed using a next-generation sequencing (NGS) workflow based on a virtual panel including 57 cardiomyopathy-related genes.
Results
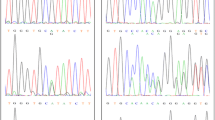
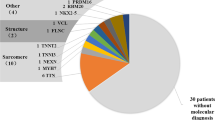
This molecular approach led to the identification of 69 cases (29.9% of the cohort) genotyped as a carrier of at least one pathogenic or likely pathogenic variant. Fourteen patients were carriers of two mutated alleles (homozygous or compound heterozygous) on the same cardiomyopathy-related gene, explaining the severe clinical disease with early-onset cardiomyopathy. Homozygous TNNI3 pathogenic variants were detected for five unrelated neonates (2.2% of the cohort), with four of them carrying the same truncating variant, i.e. p.Arg69Alafs*8.
Conclusions
Our study confirmed the importance of genetic testing in pediatric cardiomyopathies. Discovery of novel pathogenic variations is crucial for clinical management of affected families, as a positive genetic result might be used by a prospective parent for prenatal genetic testing or in the process of pre-implantation genetic diagnosis.

Similar content being viewed by others
References
McKenna WJ, Judge DP. Epidemiology of the inherited cardiomyopathies. Nat Rev Cardiol. 2021;18:22–36. https://doi.org/10.1038/s41569-020-0428-2.
Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation. 1995;92:785–9. https://doi.org/10.1161/01.cir.92.4.785.
Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis. 2006;1:27. https://doi.org/10.1186/1750-1172-1-27.
Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–47. https://doi.org/10.1038/nrcardio.2013.105.
Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–54. https://doi.org/10.1016/j.jacc.2015.01.019.
Janin A, Januel L, Cazeneuve C, Delinière A, Chevalier P, Millat G. Molecular diagnosis of inherited cardiac diseases in the era of next-generation sequencing: a single center’s experience over 5 years. Mol Diagn Ther. 2021;25:373–85. https://doi.org/10.1007/s40291-021-00530-w.
Parker LE, Landstrom AP. The clinical utility of pediatric cardiomyopathy genetic testing: from diagnosis to a precision medicine-based approach to care. Prog Pediatr Cardiol. 2021;62:101413. https://doi.org/10.1016/j.ppedcard.2021.101413.
Chanavat V, Janin A, Millat G. A fast and cost-effective molecular diagnostic tool for genetic diseases involved in sudden cardiac death. Clin Chim Acta. 2016;453:80–5. https://doi.org/10.1016/j.cca.2015.12.011.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.
Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016;99:247. https://doi.org/10.1016/j.ajhg.2016.06.001.
Millat G, Lafont E, Nony S, Rouvet I, Bozon D. Functional characterization of putative novel splicing mutations in the cardiomyopathy-causing genes. DNA Cell Biol. 2015;34:489–96. https://doi.org/10.1089/dna.2015.2842.
Kühnisch J, Herbst C, Al-Wakeel-Marquard N, Dartsch J, Holtgrewe M, Baban A, et al. Targeted panel sequencing in pediatric primary cardiomyopathy supports a critical role of TNNI3. Clin Genet. 2019;96:549–59. https://doi.org/10.1111/cge.13645.
Pezzoli L, Pezzani L, Bonanomi E, Marrone C, Scatigno A, Cereda A, et al. Not only diagnostic yield: whole-exome sequencing in infantile cardiomyopathies impacts on clinical and family management. J Cardiovasc Dev Dis. 2021;9:2. https://doi.org/10.3390/jcdd9010002.
Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, et al. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation. 2018;138:1387–98. https://doi.org/10.1161/CIRCULATIONAHA.117.033200.
Belkaya S, Kontorovich AR, Byun M, Mulero-Navarro S, Bajolle F, Cobat A, et al. Autosomal recessive cardiomyopathy presenting as acute myocarditis. J Am Coll Cardiol. 2017;69:1653–65. https://doi.org/10.1016/j.jacc.2017.01.043.
Wang J, Wang Y, Zou Y, Sun K, Wang Z, Ding H, et al. Malignant effects of multiple rare variants in sarcomere genes on the prognosis of patients with hypertrophic cardiomyopathy. Eur J Heart Fail. 2014;16:950–7. https://doi.org/10.1002/ejhf.144.
Seidel F, Holtgrewe M, Al-Wakeel-Marquard N, Opgen-Rhein B, Dartsch J, Herbst C, et al. Pathogenic variants associated with dilated cardiomyopathy predict outcome in pediatric myocarditis. Circ Genomic Precis Med. 2021;14(4):e003250. https://doi.org/10.1161/CIRCGEN.120.003250.
Olivotto I, Girolami F, Sciagrà R, Ackerman MJ, Sotgia B, Bos JM, et al. Microvascular function is selectively impaired in patients with hypertrophic cardiomyopathy and sarcomere myofilament gene mutations. J Am Coll Cardiol. 2011;58:839–48. https://doi.org/10.1016/j.jacc.2011.05.018.
Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–8. https://doi.org/10.4065/83.6.630.
Mehaney DA, Haghighi A, Embaby AK, Zeyada RA, Darwish RK, Elfeel NS, et al. Molecular analysis of dilated and left ventricular noncompaction cardiomyopathies in Egyptian children. Cardiol Young. 2021;32:295–300. https://doi.org/10.1017/S1047951121002055.
Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–35. https://doi.org/10.1093/eurheartj/ehu301.
Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. https://doi.org/10.1038/gim.2016.90.
Lu C, Wu W, Liu F, Yang K, Li J, Liu Y, et al. Molecular analysis of inherited cardiomyopathy using next generation semiconductor sequencing technologies. J Transl Med. 2018;16:241. https://doi.org/10.1186/s12967-018-1605-5.
Hu X, Li N, Xu Y, Li G, Yu T, Yao RE, et al. Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet Med. 2018;20:1045–53. https://doi.org/10.1038/gim.2017.195.
Haer-Wigman L, van der Schoot V, Feenstra I, Vulto-van Silfhout AT, Gilissen C, Brunner HG, et al. 1 in 38 individuals at risk of a dominant medically actionable disease. Eur J Hum Genet. 2019;27:325–30. https://doi.org/10.1038/s41431-018-0284-2.
Bos JM, Will ML, Gersh BJ, Kruisselbrink TM, Ommen SR, Ackerman MJ. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2014;89:727–37. https://doi.org/10.1016/j.mayocp.2014.01.025.
Kostareva A, Gudkova A, Sjöberg G, Mörner S, Semernin E, Krutikov A, et al. Deletion in TNNI3 gene is associated with restrictive cardiomyopathy. Int J Cardiol. 2009;131:410–2. https://doi.org/10.1016/j.ijcard.2007.07.108.
LaDuca H, Farwell KD, Vuong H, Lu HM, Mu W, Shahmirzadi L, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS ONE. 2017;12(2):e0170843. https://doi.org/10.1371/journal.pone.0170843.
Kaski JP, Syrris P, Burch M, Tomé-Esteban MT, Fenton M, Christiansen M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94:1478–84. https://doi.org/10.1136/hrt.2007.134684.
Jia L, Chen Y, Hao C, Guo R, Liu Y, Li W, et al. Identification of variants in TNNI3 gene in two children with restrictive cardiomyopathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2021;38:731–4. https://doi.org/10.3760/cma.j.cn511374-20200602-00406.
van den Wijngaard A, Volders P, Van Tintelen JP, Jongbloed JD, van den Berg MP, Lekanne Deprez RH, et al. Recurrent and founder mutations in the Netherlands: cardiac Troponin I (TNNI3) gene mutations as a cause of severe forms of hypertrophic and restrictive cardiomyopathy. Neth Heart J. 2011;19:344–51. https://doi.org/10.1007/s12471-011-0135-z.
Burns C, Bagnall RD, Lam L, Semsarian C, Ingles J. Multiple gene variants in hypertrophic cardiomyopathy in the era of next-generation sequencing. Circ Cardiovasc Genet. 2017;10(4):e001666. https://doi.org/10.1161/CIRCGENETICS.116.001666.
Shah S, Yogasundaram H, Basu R, Wang F, Paterson DI, Alastalo TP, et al. Novel Dominant-Negative Mutation in Cardiac Troponin I Causes Severe Restrictive Cardiomyopathy. Circ Heart Fail. 2017;10(2):e003820. https://doi.org/10.1161/CIRCHEARTFAILURE.116.003820.
Mörner S, Richard P, Kazzam E, Hainque B, Schwartz K, Waldenström A. Deletion in the cardiac troponin I gene in a family from northern Sweden with hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:521–5. https://doi.org/10.1006/jmcc.1999.1099.
Cheng Y, Regnier M. Cardiac troponin structure-function and the influence of hypertrophic cardiomyopathy associated mutations on modulation of contractility. Arch Biochem Biophys. 2016;601:11–21. https://doi.org/10.1016/j.abb.2016.02.004.
Acknowledgements
The authors are grateful to the patients and families, as well as to all colleagues who, over many years, provided them with biological samples and with invaluable clinical information.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Molecular diagnosis was supported by Hospices Civils de Lyon (France).
Conflict of interest
Alexandre Janin, Thomas Perouse de Montclos, Cécile Cazeneuve, Rajae El-Malti, Elodie Morel, Antoine Delinière, Philippe Chevalier, and Gilles Millat are employees of Hospices Civils de Lyon (HCL), France. Karine Nguyen and Emilie Consolino are employees of Assistance Publique Hôpitaux Marseille (APHM), France. Gwenael Nadeau and Gaelle Rey are employees of Metropole Savoie Hospital Center at Chambéry, France. Océane Bouchot is employee of Centre Hospitalier Annecy, Genevois, France. Patricia Blanchet is employee of CHU de Montpellier, France. Quentin Sabbagh is an employee of Département de Génétique Médicale, CHU de Montpellier, Montpellier, France. None of the authors have any potential conflicts of interest to declare.
Informed consent and ethical approval
This study was conducted in accordance with the principles of the Declaration of Helsinki and informed consent was obtained for all cases.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
Authors’ contributions
AJ, CC, and GM are Molecular Biologists involved in all molecular diagnosis steps (from blood sample to clinical reports). TPM, AD, PC and OB are Cardiologists involved in the clinical follow-up of the different families reported in this study. REM and EM are Clinical Research Assistants at the National Reference Center of Inherited Cardiac Diseases in Lyon. KN, EC, GN, GR, QS and PB are medical geneticists involved in genetic follow-up of the different families reported in this study (genetic counseling).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Janin, A., Perouse de Montclos, T., Nguyen, K. et al. Molecular Diagnosis of Primary Cardiomyopathy in 231 Unrelated Pediatric Cases by Panel-Based Next-Generation Sequencing: A Major Focus on Five Carriers of Biallelic TNNI3 Pathogenic Variants. Mol Diagn Ther 26, 551–560 (2022). https://doi.org/10.1007/s40291-022-00604-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-022-00604-3