Abstract
Background and Objective
Membrane solute carrier transporters play an important role in the transport of a wide spectrum of substrates including anticancer drugs and cancer-related physiological substrates. This study aimed to assess the prognostic relevance of gene expression and genetic variability of selected solute carrier transporters in breast cancer.
Methods
Gene expression was determined by quantitative real-time polymerase chain reaction. All SLC46A1 and SLCO1A2 exons and surrounding non-coding sequences in DNA extracted from the blood of patients with breast cancer (exploratory phase) were analyzed by next-generation sequencing technology. Common variants (minor allele frequency ≥ 5%) with in silico-predicted functional relevance were further analyzed in a large cohort of patients with breast cancer (n = 815) and their prognostic and predictive potential was estimated (validation phase).
Results
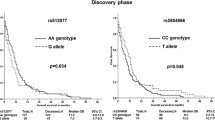
A gene expression and bioinformatics analysis suggested SLC46A1 and SLCO1A2 to play a putative role in the prognosis of patients with breast cancer. In total, 135 genetic variants (20 novel) were identified in both genes in the exploratory phase. Of these variants, 130 were non-coding, three missense, and two synonymous. One common variant in SLCO1A2 and four variants in SLC46A1 were predicted to be pathogenic by in silico programs and subsequently validated. A SLC46A1 haplotype block composed of rs2239911-rs2239910-rs8079943 was significantly associated with ERBB2/HER2 status and disease-free survival of hormonally treated patients.
Conclusions
This study revealed the prognostic value of a SLC46A1 haplotype block for breast cancer that should be further studied.




Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Videira M, Reis RL, Brito MA. Deconstructing breast cancer cell biology and the mechanisms of multidrug resistance. Biochim Biophys Acta. 2014;1846(2):312–25.
El-Gebali S, Bentz S, Hediger M, Anderle P. Solute carriers (SLCs) in cancer. Mol Aspects Med. 2013;34(2–3):719–34.
Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–34.
Lemstrova R, Soucek P, Melichar B, Mohelnikova-Duchonova B. Role of solute carrier transporters in pancreatic cancer: a review. Pharmacogenomics. 2014;15(8):1133–45.
Mohelnikova-Duchonova B, Melichar B, Soucek P. FOLFOX/FOLFIRI pharmacogenetics: the call for a personalized approach in colorectal cancer therapy. World J Gastroenterol. 2014;20(30):10316–30.
Hlavata I, Mohelnikova-Duchonova B, Vaclavikova R, Liska V, Pitule P, Novak P, et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis. 2012;27(2):187–96.
Hlavac V, Brynychova V, Vaclavikova R, Ehrlichova M, Vrana D, Pecha V, et al. The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics. 2013;14(5):515–29.
Elsnerova K, Mohelnikova-Duchonova B, Cerovska E, Ehrlichova M, Gut I, Rob L, et al. Gene expression of membrane transporters: importance for prognosis and progression of ovarian carcinoma. Oncol Rep. 2016;35(4):2159–70.
Mohelnikova-Duchonova B, Brynychova V, Hlavac V, Kocik M, Oliverius M, Hlavsa J, et al. The association between the expression of solute carrier transporters and the prognosis of pancreatic cancer. Cancer Chemother Pharmacol. 2013;72(3):669–82.
Mohelnikova-Duchonova B, Brynychova V, Oliverius M, Honsova E, Kala Z, Muckova K, et al. Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas. 2013;42(4):707–16.
César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, et al. A call for systematic research on solute carriers. Cell. 2015;162(3):478–87.
Zemanek T, Melichar B, Lovecek M, Soucek P, Mohelnikova-Duchonova B. Biomarkers and pathways of chemoresistance and chemosensitivity for personalized treatment of pancreatic adenocarcinoma. Pharmacogenomics. 2019;20(2):113–27.
Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflug Arch. 2004;447:735–43.
Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflug Arch. 2004;447:728–34.
Qiu A, Jansen M, Sakaris A, Min SH, Chattopadhyay S, Tsai E, et al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–28.
Sprowl JA, Mikkelsen TS, Giovinazzo H, Sparreboom A. Contribution of tumoral and host solute carriers to clinical drug response. Drug Resist Updates. 2012;15:5–20.
Angelini S, Pantaleo MA, Ravegnini G, Zenesini C, Cavrini G, Nannini M, et al. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharm Res. 2013;68:1–6.
Nakanishi T, Tamai I. Putative roles of organic anion transporting polypeptides (OATPs) in cell survival and progression of human cancers. Biopharm Drug Dispos. 2014;35(8):463–84.
Sutherland R, Meeson A, Lowes S. Solute transporters and malignancy: establishing the role of uptake transporters in breast cancer and breast cancer metastasis. Cancer Metast Rev. 2020;39(3):919–32.
Hubackova M, Vaclavikova R, Ehrlichova M, Mrhalova M, Kodet R, Kubackova K, et al. Association of superoxide dismutases and NAD(P)H quinone oxidoreductases with prognosis of patients with breast carcinomas. Int J Cancer. 2012;130:338–48.
Hlavac V, Kovacova M, Elsnerova K, Brynychova V, Kozevnikovova R, Raus K, et al. Use of germline genetic variability for prediction of chemoresistance and prognosis of breast cancer patients. Cancers. 2018;10:511.
Therasse P, Eisenhauer EA, Verweij J. RECIST revisited: a review of validation studies on tumour assessment. Eur J Cancer. 2006;42(8):1031–9.
Topic E, Gluhak J. Isolation of restrictible DNA. Eur J Clin Chem Clin Biochem. 1991;29(5):327–30.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22.
Vaclavikova R, Klajic J, Brynychova V, Elsnerova K, Alnaes GIG, Tost J, et al. Development of high resolution melting analysis for ABCB1 promoter methylation: clinical consequences in breast and ovarian carcinoma. Oncol Rep. 2019;42(2):763–74.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1.
Koch A, De Meyer T, Jeschke J, Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genom. 2015;16:636.
Nagy A, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43(1110):11.10.1-33.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9.
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11(1):1–9.
Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300.
Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharm Ther. 2012;92(4):414–7.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–6.
Jarman EJ, Ward C, Turnbull AK, Martinez-Perez C, Meehan J, Xintaropoulou C, et al. HER2 regulates HIF-2alpha and drives an increased hypoxic response in breast cancer. Breast Cancer Res. 2019;21(1):10.
Clifford AJ, Rincon G, Owens JE, Medrano JF, Moshfegh AJ, Baer DJ, et al. Single nucleotide polymorphisms in CETP, SLC46A1, SLC19A1, CD36, BCMO1, APOA5, and ABCA1 are significant predictors of plasma HDL in healthy adults. Lipids Health Dis. 2013;12:66.
Lima A, Bernardes M, Azevedo R, Monteiro J, Sousa H, Medeiros R, et al. SLC19A1, SLC46A1 and SLCO1B1 polymorphisms as predictors of methotrexate-related toxicity in Portuguese rheumatoid arthritis patients. Toxicol Sci. 2014;142(1):196–209.
Huang T, Tucker KL, Lee YC, Crott JW, Parnell LD, Shen J, et al. Interactions between genetic variants of folate metabolism genes and lifestyle affect plasma homocysteine concentrations in the Boston Puerto Rican population. Public Health Nutr. 2011;14(10):1805–12.
Kelley KM, Rowan BG, Ratnam M. Modulation of the folate receptor alpha gene by the estrogen receptor: mechanism and implications in tumor targeting. Cancer Res. 2003;63(11):2820–8.
Price RJ, Lillycrop KA, Burdge GC. Folic acid induces cell type-specific changes in the transcriptome of breast cancer cell lines: a proof-of-concept study. J Nutr Sci. 2016;5:e17.
Zhou F, Zhu L, Wang K, Murray M. Recent advance in the pharmacogenomics of human solute carrier transporters (SLCs) in drug disposition. Adv Drug Deliv Rev. 2017;116:21–36.
Callens C, Debled M, Delord M, Turbiez-Stalain I, Veyret C, Bièche I, Brain E. High-throughput pharmacogenetics identifies SLCO1A2 polymorphisms as candidates to elucidate the risk of febrile neutropenia in the breast cancer RAPP-01 trial. Breast Cancer Res Treat. 2015;153(2):383–9.
Lee W, Glaeser H, Smith LH, Roberts RL, Moeckel GW, Gervasini G, et al. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280(10):9610–7.
Badagnani I, Castro RA, Taylor TR, Brett CM, Huang CC, Stryke D, et al. Interaction of methotrexate with organic-anion transporting polypeptide 1A2 and its genetic variants. J Pharmacol Exp Ther. 2006;318(2):521–9.
Eechoute K, Franke RM, Loos WJ, Scherkenbach LA, Boere I, Verweij J, et al. Environmental and genetic factors affecting transport of imatinib by OATP1A2. Clin Pharmacol Ther. 2011;89(6):816–20.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the Ministry of Health of the Czech Republic, (grant no. 17-28470A to P.S.), the National Center of Medical Genomics (project no. CZ.02.1.01/0.0/0.0/16_013/0001634 to V.H.), the Czech Ministry of Education, Youth and Sports INTER-COST project no. LTC19020 (COST Action CA17104 STRATAGEM to R.V.), and the Charles University project “Center of clinical and experimental liver surgery” (no. UNCE/MED/006 to P.D.).
Conflicts of Interest/Competing interests
Viktor Hlavac, Radka Vaclavikova, Veronika Brynychova, Pavel Dvorak, Katerina Elsnerova, Renata Kozevnikovova, Karel Raus, Katerina Kopeckova, Sona Mestakova, David Vrana, Jiri Gatek, and Pavel Soucek have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Procedures performed in the present study were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Study protocol was approved by the Ethical Commission of the National Institute of Public Health in Prague (approval nos. 9799-4 and 15-25618A).
Consent to participate
All patients were informed about the study and those who agreed and signed an informed consent participated in the study.
Consent for publication
Not applicable.
Availability of data and material
The sequencing data that support the findings of this study are openly available in Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession no. PRJNA510917.
Code availability
Not applicable.
Authors’ contributions
Study conceptualization and interpretation of results (VH, PS), experimental work and evaluation of results (VH, RV, VB, KE), bioinformatics (PD), collection of clinical samples and patient follow-up and data management (RK, KR, KK, SM, DV, JG), and drafting the manuscript (all authors).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hlavac, V., Vaclavikova, R., Brynychova, V. et al. SLC46A1 Haplotype with Predicted Functional Impact has Prognostic Value in Breast Carcinoma. Mol Diagn Ther 25, 99–110 (2021). https://doi.org/10.1007/s40291-020-00506-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00506-2