Abstract
Background
The clinicopathological features and genomic rearrangements of anaplastic lymphoma kinase (ALK) fusion cases have not been fully identified.
Objective
Our objective was to explore the status of ALK in non-small-cell lung cancer (NSCLC) specimens, to explore the relationships between ALK status and clinicopathological features and to identify genomic rearrangements via capture-based next-generation sequencing (NGS).
Methods
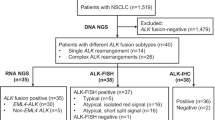
We tested 9889 NSCLC specimens for ALK status using the Ventana anti-ALK (D5F3) antibody. Clinicopathological features were analyzed and genomic rearrangements identified using capture-based NGS in 76 ALK-positive cases.
Results
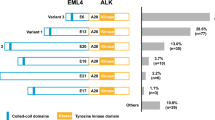
In total, 485 specimens (4.90%) tested positive for ALK. The positivity rate was higher for adenocarcinoma samples than for non-adenocarcinoma samples (6.03 vs. 1.47%; p < 0.001) and for biopsies/cell blocks than for surgical specimens (7.00 vs. 4.16%; p < 0.001). Patient age, patient sex, specimen type, specimen histotype, and patient smoking history were all significantly correlated with ALK status. Genomic rearrangements were detected in 98.68% (75/76) of the ALK-positive samples; 89.33% (67/75) carried the canonical EML4-ALK, and the remaining samples carried only noncanonical ALK rearrangements. Four of these noncanonical ALK fusion samples were identified as carrying EML4-ALK transcripts at the RNA level. A novel fusion variant, SRD5A2-ALK, was revealed.
Conclusions
Younger patients with NSCLC, especially those aged < 30 years, were more likely to test positive for ALK. Positive ALK test results were more common in patients with invasive mucinous adenocarcinoma and solid-predominant invasive adenocarcinoma than in patients with other histotypes. Samples that carried only noncanonical ALK rearrangements may also have carried the canonical EML4-ALK, which was not detected by capture-based NGS. EML4-ALK transcripts might result from rare splicing mechanisms without genomic rearrangements.



Similar content being viewed by others
References
Cui JJ, Tran-Dubé M, Shen H, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem. 2011;54(18):6342–63. https://doi.org/10.1021/jm2007613.
Wynes MW, Sholl LM, Dietel M, et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol. 2014;9(5):631–8. https://doi.org/10.1097/JTO.0000000000000115.
von Laffert M, Warth A, Penzel R, et al. Multicenter immunohistochemical ALK-testing of non-small-cell lung cancer shows high concordance after harmonization of techniques and interpretation criteria. J Thorac Oncol. 2014;9(11):1685–92. https://doi.org/10.1097/JTO.0000000000000332.
Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. https://doi.org/10.1038/nature05945.
Sasaki T, Rodig S J, Chirieac L R, et al. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46(10):1773–1780. https://doi.org/10.1016/j.ejca.2010.04.002.
Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21(16):3631–9. https://doi.org/10.1158/1078-0432.CCR-14-2683.
Takeuchi K, Togashi Y, Kamihara Y, et al. Prospective and clinical validation of ALK immunohistochemistry: results from the phase I/II study of alectinib for ALK-positive lung cancer (AF-001JP study). Ann Oncol. 2016;27(1):185–92. https://doi.org/10.1093/annonc/mdv501.
Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22(4):508–15. https://doi.org/10.1038/modpathol.2009.2.
Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–53. https://doi.org/10.1200/JCO.2009.22.6993.
Hong S, Fang W, Hu Z, et al. A large-scale cross-sectional study of ALK rearrangements and EGFR mutations in non-small-cell lung cancer in Chinese Han population. Sci Rep. 2014;4:7268. https://doi.org/10.1038/srep07268.
Jokoji R, Yamasaki T, Minami S, et al. Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol. 2010;63(12):1066–70. https://doi.org/10.1136/jcp.2010.081166.
Sakairi Y, Nakajima T, Yasufuku K, et al. EML4-ALK fusion gene assessment using metastatic lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration. Clin Cancer Res. 2010;16(20):4938–45. https://doi.org/10.1158/1078-0432.CCR-10-0099.
Wang W, Song Z, Zhong Y. Response to crizotinib in a squamous cell lung carcinoma patient harbouring echinoderm microtubule-associated protein-like 4-anaplastic lymphoma translocation: a case report. Thorac Cancer. 2016;7(3):355–7. https://doi.org/10.1111/1759-7714.12298.
Zhao W, Choi YL, Song JY, et al. ALK, ROS1 and RET rearrangements in lung squamous cell carcinoma are very rare. Lung Cancer. 2016;94:22–7. https://doi.org/10.1016/j.lungcan.2016.01.011.
Crystal AS, Shaw AT. New targets in advanced NSCLC: EML4-ALK. Clin Adv Hematol Oncol. 2011;9(3):207–14.
Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. https://doi.org/10.1038/nrc3580.
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. https://doi.org/10.1016/j.cell.2007.11.025.
Rosenbaum JN, Bloom R, Forys JT, et al. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Modern Pathology. 2018;31(5):791–808. https://doi.org/10.1038/modpathol.2017.181.
Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. https://doi.org/10.1158/1078-0432.CCR-08-3248.
Fang DD, Zhang B, Gu Q, et al. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol. 2014;9:285–94. https://doi.org/10.1097/JTO.0000000000000087.
Qing TIAN, Wen-Jing DENG, Zong-Wei LI. Identification of a novel crizotinib-sensitive BCL11A-ALK gene fusion in a non-small cell lung cancer patient. European Respiratory Journal. 2017;49(4):1602149. https://doi.org/10.1183/13993003.02149-2016.
Jividen K, Li H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes, Chromosomes and Cancer. 2014;53(12):963–71. https://doi.org/10.1002/gcc.22207.
Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–94. https://doi.org/10.1056/NEJMoa1214886.
Salido M, Pijuan L, Martinez-Aviles L, et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol. 2011;6:21–7. https://doi.org/10.1097/JTO.0b013e3181fb7cd6.
Wiesner T, Lee W, Obenauf AC, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–7. https://doi.org/10.1038/nature15258.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was funded by the Shanghai Municipal Commission of Health and Family Planning (Grant number 2018ZHYL0213), the Interdisciplinary Program of Shanghai Jiao Tong University (Grant number ZH2018QNA66) and the Shanghai Chest Hospital Projects for Development of Science and Technology (Grant number YZ2015-ZX).
Conflict of Interest
Ruiying Zhao, Jie Zhang, Yuchen Han, Jinchen Shao, Lei Zhu, Chan Xiang, Qing Zhang, Haohua Teng, Gang Qin, Lanxiang Zhao, Min Ye, Jikai Zhao, and Wenjie Ding have no conflicts of interest that are directly relevant to the content of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, R., Zhang, J., Han, Y. et al. Clinicopathological Features of ALK Expression in 9889 Cases of Non-small-Cell Lung Cancer and Genomic Rearrangements Identified by Capture-Based Next-Generation Sequencing: A Chinese Retrospective Analysis. Mol Diagn Ther 23, 395–405 (2019). https://doi.org/10.1007/s40291-019-00389-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00389-y