Abstract
Advanced health economic analysis techniques currently performed in Microsoft Excel, such as incorporating heterogeneity, time-dependent transitions and a value of information analysis, can be easily transferred to R. Often the outputs of survival analyses (such as Weibull regression models) will estimate the impacts of correlated patient characteristics on patient outcomes, and are utilised directly as inputs for health economic decision models. This tutorial provides a step-by-step guide of how to conduct such analyses with a Markov model developed in R, and offers a comparison with established analyses performed in Microsoft Excel. This is done through the conversion of a previously published Microsoft Excel case study of a hip replacement surgery cost-effectiveness model. We hope that this paper can act as a facilitator in switching decision models from Microsoft Excel to R for complex health economic analyses, providing open-access code and data, suitable for future adaptation.
Similar content being viewed by others
First tutorial for linking survival analyses to Markov models and performing sensitivity analyses (including a value of information analysis) in R. |
Intended for users of Microsoft Excel with downloadable resources across both types of software, with a practical example for total hip replacement prosthesis strategies. |
Provides adaptable open-access resources to be used as frameworks for future health economic evaluation models in R. |
1 Introduction
The benefits of utilising R for health economic evaluations are becoming well documented [1,2,3]. Whilst Microsoft (MS) Excel and TreeAge are visual graphical user interfaces and therefore useful software for learning purposes, R (alongside other programming language-based software such as MATLAB) has higher efficiency, transparency and adaptability in comparison [1–3].
The foundation of many such health economic evaluations is often the Markov model. Markov models can quantify the impact of interventions on transitions between health states, as well as the costs and outcomes associated with the differing course of actions. Intervention impacts on health outcomes, conditional on patient characteristics, can be quantified through standard survival analysis techniques. Subsequently, these impacts can be fed through Markov models to appropriately account for heterogeneity across subpopulations of interest. Additionally, decomposition techniques can be utilised to allow for covariance to be maintained during probabilistic sensitivity analyses.
Previous health economic evaluation tutorials for R generally run through how to create deterministic and probabilistic Markov models in R [4]. However, a comparison of more advanced modelling techniques, such as modelling heterogeneity through the inclusion of survival analysis results whilst conducting value of information (VOI) analyses in R compared to MS Excel, has yet to be done. This tutorial first introduces a case study of hip replacement surgery, for which an MS Excel model has been published [5]. This case study is then used to demonstrate how to integrate survival analyses within sensitivity analyses using R instead of MS Excel. This is then followed by instructions on how to conduct analyses for the expected value of perfect information (EVPI) and the expected value of partially perfect information (EVPPI), also known as the expected value of perfect parameter information, within R. We outline how to conduct these analyses using mainly base R functions. By focusing on using basic R functions, rather than specific health economics R packages, it reduces reliance on “black boxes” and increases the potential for adaptability to suit need.
2 General Tips
2.1 Setting Up Projects
Throughout this tutorial code, data available from the GitHub repository are cited [6]. Once the folder has been downloaded (which can be done through the “code” button) or linked via another interface, such as RStudio, you can run the R project. Clicking on the Git “R Project” item automatically sets your working directory to be the equivalent to where the project is based, allowing you to read in data also within that folder. There are many blogs and guides on how to link Git and RStudio using R projects [7]. The RProject is the health economic evaluation model. The folder containing the RProject files acts as the “MS Excel file”, where the separate csv files and R scripts within that folder are similar to having different MS Excel sheets within the file, housing different data tables or analysis functions for the model.
2.2 Reading in Data
In this tutorial, we read in data files that contain life table data and outputs of survival analyses. For csv files, base R (i.e. R “as is”) allows you to read in data using the ‘read.csv()’ function, where the file path relative to the working directory can be specified within the brackets. For example ‘read.csv("life-table.csv", header=TRUE)’ reads in the life table csv data in the working directory, specifying that the first row of the csv file is the header row. For other types of data files, there are often already packages that deal with directly importing such data (e.g. the ‘xlsx’ package importing MS Excel ‘.xls’ and ‘.xlsx’ files [8]).
2.3 Graphics Packages
This tutorial uses base R wherever possible, reducing reliance on packages, and often allows for better understanding for each step. However, we do use the reshape2 [9] and ggplot2 [10] packages. reshape2 allows for easy data manipulation (e.g. converting data between long and wide formats), whilst ggplot2 allows users to create attractive plots and diagrams that suit need (e.g. plotting multiple variables on multiple panels).
3 Total Hip Replacement: A Case Study for an Advanced Health Economic Evaluation
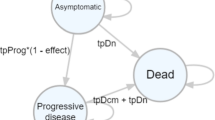
Cost utility models for the total hip replacement case study are available online, performed in MS Excel [11, 12]. These are a simplified version of a previously published economic model, developed for education purposes [5]. This tutorial will use the same case study when demonstrating how to utilise R when conducting more advanced health economic modelling. Within this example, a new procedure that reduces the risk of revision surgery in a cohort of patients undergoing a primary total hip replacement (THR) procedure is compared to standard treatment. Potential health states and state transitions are displayed in Fig. 1.
Total hip replacement (THR) decision model schematic. Health states of the model are represented by ovals, transitions between health states are represented by arrows. [C] represents a collapsed node of the decision tree in which the Markov model [M] is repeated. *Procedures are represented by rectangles whereby primary hip replacement is either the new or standard procedure depending on the respective branch on the decision tree
All of the R relevant materials used in this tutorial can be found within a corresponding GitHub repository [6]. The corresponding MS Excel files are available for download within this repository; or originally from the Briggs et al. corresponding webpage [11]. Specifically, “THR_Model.R” and “THR_Model_VOI.R” scripts are equivalent to “Ex57sol.xls” and “Ex66bsol.xls”, respectively. The Markov model process or probabilistic analysis will not be described in detail here, as these are covered in detail elsewhere [4].
In this tutorial, we follow the structure outlined in Fig. 2, referring to the stated subsection headings and relating these to equivalent MS Excel processes. This structure can be utilised in other models outside of the case study.
3.1 Set-Up Model Structure
In the MS Excel model, there are different worksheets that house inputs, intermediate values and outputs, including “Parameters” listing the main parameters of the model and “Standard” listing the health states and tracing the cohort across these health states over time for a standard prosthesis (see in Fig. 1 for health states included). Information from different sheets is then combined to produce outputs presented on the analysis sheet. In R, we focus on one script that follows the sections described in Fig. 2 (see ‘THR_Model.R’ [6]) to define inputs and produce outputs. Numbered sections refer to subheadings within the relevant R script.
To set up the model in R, we first need to load packages, which will be used later for plotting data. However, it is good practice to group and load all necessary packages at the top of the script, avoiding potential issues when running parts of your code [13]. We load reshape2 and ggplot2, which are needed to plot the outputs [9, 10].
There are two ways in which age and sex influence transitions within this case study, with mean ages of 40, 60 and 80 years for male and female individuals of interest. The first is through background mortality and the second through the impact on the risk of failure of primary THR. External data in the form of life tables (“life-table.csv”) and a parametric Weibull regression analysis (“hazardfunction.csv” and “cov55.csv” containing coefficient and covariance data, respectively) need to be fed into the model. In MS Excel, these are simply added/viewed as worksheets. It is also good practice to read in data utilised within the R script at the beginning, after packages. By having a dedicated section at the beginning of your script dedicated to external code and data, any errors due to reading in such information are detected early and allow for easy testing of other portions of code reliant on these external factors.
A benefit of using R, over MS Excel’s RAND-type functions, is that you can “set the seed” easily within your script so that when you draw from a sample (random sampling), R returns the same values every time. This is achieved by entering set.seed(#), where # is set to an integer of your choice (e.g. 100) and corresponds with the draw you will obtain. By setting the seed, you can ensure consistency in result reporting and model checking.
The THR model script therefore begins with:
-
i.
Loading libraries, such as ggplot2 [10]:
$$if\left( {!require\left( {ggplot2} \right)} \right) install.packages\left( {{}^{^{\prime}}ggplot2^{^{\prime}} } \right)$$$$library\left( {ggplot2} \right),$$where the initial “if” statement checks to see if the user has the package installed before calling it to be used, and if not, installs the package.
-
ii.
Reading in data files, such as the life tables:
$$life.tables~ <\! - ~read.csv\left( {\hbox{``}life - table.csv\hbox{''} ,~header = TRUE} \right)$$ -
iii.
Setting the seed:
$$set.seed\left( {1234} \right)$$
Structural parameters then need to be defined, through declaring state names ('state.names') and numbers ('n.states'), initial cohort distribution across health states ('seed'), number of cycles (in the case study, 'cycles' are set to 60 and 'cycle.v' being a vector from 1 to 60), discount rates for costs (cDR) and outcomes (oDR), using assignments performed in a previous tutorial [4]. Discount rates can be included in models in various ways. One approach is to define a vector of discount weights that can be easily multiplied by resulting outcome vectors or matrices over time (see Fig. 3a for code and Fig. 3b for output examples). A discount rate of 1.5% for outcomes and 6% for costs are used in the case study.
3.2 Define Deterministic Parameters
Focusing on setting the deterministic model parameters, the shape and scale parameters for those to be used in probabilistic analyses later, means naming and assigning values to variables and health states as seen in Table 1.
Table 1 shows that only four variables are deterministic; the cost of primary surgery and successful surgery (set to zero as these are the same values across comparators and we are interested in incremental analyses) and the cost of a standard prosthesis (£394) and a new prosthesis (£579). The remainder are probabilistic, with distributions presented in Table 1.
3.3 Defining the Shape and Scale Values for Probabilistic Parameters
The parameter values in the probabilistic sensitivity analysis (PSA) distributions in Table 1 can be determined by the mean and standard error of the parameters if known. For example, for the cost of revision where the mean cost is £5294, and the standard error is £1487:
This process is performed for the other probabilistic values, apart from for the risk of revision and the risk of death. The former first needs to integrate the results of a survival analysis. Once these values are estimated, they are stored in a list ('params'), so that they can be passed to the model function, later on.
3.4 Defining Hazard Coefficient and Covariance Values
To utilise the Weibull regression results, we first save the coefficient values (which represents the hazard ratios for each coefficient) within the list of parameter values (‘params’). This is stored as a vector within the ‘params’ list, and includes the constant, age, sex and new prosthesis coefficients. We can also save the covariance matrix associated with this particular survival analysis:
This is similar to using the Name functionality within MS Excel (under “Formula” and “Define Name”) to label parameter values and/or tables within sheets, which can then be utilised in formulae instead of referring to the cell number itself. An advantage of using R for this process over MS Excel is, as it is a script language, users can see which ‘named values’ come from which source, and in what order they are assigned and used, instead of having various sheets for which the inter-sheet dependency can be opaque, unless thoroughly annotated.
3.5 Sampling
To incorporate heterogeneity in the risks of revision and death due to age and sex, whilst performing sensitivity analyses, a function needs to be created to produce a list (‘sample.output’). This list houses samples for each parameter/parameter group of interest. This is equivalent to sampling every parameter value, for every iteration of the PSA, for all of the parameters included in the PSA, and storing in an Excel SheetFootnote 1 so that it is clear which values are being used in each PSA simulation. The function allows users to specify age (as a numeric) and sex (as a dummy variable that is 0 for female individuals and 1 for male individuals), the list of parameters and the number of simulation runs:
First, the covariance matrix stored in the section above can undergo a Cholesky decomposition to allow for the generation of correlated variables. For a further description of the theory behind this process, refer to Chapter 4, Briggs et al. [12]. Step-by-step calculations for the Cholesky decomposition in R are available in the Electronic Supplementary Material (ESM) to show the workings.Footnote 2 However, we use the handy ‘t()’ and ‘chol() functions available in base R (i.e. no further packages are needed to perform these operations). The transpose function (‘t()’) is needed as the Cholesky decomposition function (‘chol()’) returns the Upper Triangular Decomposition, whereas for our purposes we want the Lower Triangular Decomposition. We therefore transpose our input, run ‘chol()’, and transpose the output to produce a matrix named ‘cholm’:
The ‘cholm’ matrix can then be used in a sampling function to generate random numbers that follow the same covariance as indicated by the survival analysis. First, an empty matrix (‘temp.values’) where there are five independent standard normal variates (representing the coefficients for the five survival analysis variables ‘lngamma, cons, age, male and NP1’) for each simulation is created. For each simulation run, another matrix ('Tz') can be created that is the product of the decomposition matrix (‘cholm’) and the generated ‘temp.values’ matrix. This can then be added to the mean coefficient values (‘params$coeff’). The resulting matrix (‘coeff.table’) gives five variables that are correlated in line with the survival analyses results. An exponentiation process to correctly interpret the results of the Weibull model is then performed, as depicted in Fig. 4a. This is based on the following formulae [12], where the time-dependent transition probability per time-step (‘tp(t)’) is given by:
where lambda and gamma values are the shape and scale parameters in the Weibull distribution (see Fig. 4b).
Other probabilistic cost parameters are sampled according to the distributions outlined in Table 1. To incorporate background mortality based on life tables we need two data points: age at that cycle and mortality risk at that specific age. A process on how to do this has been previously described [4], with an additional method of doing this given in the ESM.
3.6 Total Hip Replacement Model Function
Once we have the samples for relevant parameters/parameter groups, they can be fed through the main model function (model.THR(), see ESM). The main model processes are similar to those previously described [4, 14]). To integrate the heterogeneous risks of revision and mortality within the function, simply update the transitions within the transition arrays (such as ‘tm.SP0’, which is a transition array for standard procedure where the third dimension represents each cycle) as shown in Fig. 5a, b. In this example, we set age to 60 years and male sex to 0, the code sits within a function and thus the outputs printed here are exemplary of what occurs within the function.
The model function returns a vector of values containing the costs and quality-adjusted life-years of the standard and new prosthesis procedures and the incremental cost-effectiveness ratio value of the comparison.
3.7 Running the Simulations, Estimating NMB and Performing Subgroup Analyses Across All Subgroups
Once the parameters have been sampled and the model function defined, it is a case of creating a blank data frame, which has columns representing model outputs, and rows representing the number of simulations (like having a blank sheet where each row gets filled with each simulation result in MS Excel). This can then be filled utilising a for loop, where each value of the sample outputs, created in section (5), is utilised within the model function (see Table 1 of the ESM for an example of the resulting output “simulation.outputs”).
The incremental costs and effects for each simulation run can be transformed into net monetary benefit (NMB) using willingness-to-pay thresholds (WTPs), compared to zero (with NMB >0 flagged as cost effective) and the probability of the new procedure being cost effective calculated by averaging over all simulations the number of times the procedure is deemed cost effective. This can all be wrapped into a function (‘p.CE’):
where the function is run across different WTP values and the related probability of cost effectiveness calculated, both values are then stored in a data frame. The processes outlined above can then be performed on specific age and sex subgroups, and stored within lists and/or arrays. The resulting outputs can be plotted, utilising ‘ggplot()’ allows graphs to be easily altered to suit users’ needs [10], see the ESM for the code and plots related to graphical outputs.
4 VOI Analysis
In this section, we demonstrate the steps involved in the VOI analyses, with comparisons between R code and MS Excel directly (i.e. the cells within the spreadsheet), or between R code and MS Excel’s Visual Basic for Applications code. There are two key methods for quantifying the VOI for parameter uncertainty: by calculating the EVPI and by calculating the EVPPI (for more information on the theory behind these methods, see Briggs et al. [12]). The structure of the VOI R script is shown in Fig. 6.
4.1 Set Up Model Script and Source THR Model Functions
The ‘THR_Model_VOI.R’ script is an extension of the ‘THR_Model.R script’ [6], using the same sampled data and main model function. A benefit of using R over MS Excel, is the ability to easily and efficiently source data and functions from other scripts within projects, simply by using ‘source("THR_Model.R")’ at the beginning of the VOI model script.
4.2 Set VOI Population Parameters
When we estimate the VOI, this is often done for one individual in the model. Of course, there will be many people eligible for a particular treatment, each year. The EVPI and EVPPI for an individual can be multiplied by the effective population; the number of eligible patients per annum and the expected lifetime of the technology. For the hip replacement example, we assume an effective technology life of 10 years with 40,000 new patients eligible for treatment each year. A 6% discount rate is used on the effective population, over a 10-year time horizon. The total (discounted) effective population is 312,067 (the sum of the annual discounted population over 10 years, thus e.g., if the discount rate was 0% this would be 400,000).
4.3 EVPI at the Population Level
The EVPI represents the value of eliminating the uncertainty in the model parameters, and thereby assuring that with perfect information, the correct decision is made. In contrast, when a decision is made with the current information, there may still be uncertainty around the cost effectiveness in probabilistic analyses, meaning there is a possibility that the wrong decision is made, which would result in a loss of health benefits and/or resources. A full comparison between the R and the MS Excel Visual Basic for Applications codes used for the case study, with annotated comparisons, is available in the ESM, and the full EVPPI R code is available in the ESM. The corresponding MS Excel workbook (“ExcelVersion_Ex66bsol.xlsx”) is available to download [6]).
The EVPI is estimated using the results of the PSA. The costs and effects of each PSA simulation are converted into NMB, for any given WTP value. Then, the mean NMB is taken for the two treatments, with the highest being the treatment of choice. Across each individual simulation, the highest NMB possible from either treatment is also recorded, which represents the correct decision being made in each simulation. The average of all these values gives the NMB under perfect information, which is then multiplied by the effective population.
This can all be wrapped into a function in R (‘est.EVPI.pop’), which takes the PSA results (‘simulation.results’) and creates a data frame of NMB values (‘nmb.table’) for the two treatments, across all simulations. An example of the calculations performed within the EVPI function is shown in Table 2. For example, at a WTP threshold of £2500 per quality-adjusted life-year gained, the highest NMB under current information is £36,117 (for the new prosthesis). In contrast, under perfect information, the NMB is £36,135. This gives an EVPI per person of £18.36, which multiplied by the effective population gives an EVPI of £5,730,624.
4.4 Set Up the EVPPI Inner and Outer Loop Framework and NMB Function
The population EVPI is an upper limit on returns to future research. However, of crucial importance is which particular parameters (or groups of related parameters) are most important in terms of having the greatest VOI. The EVPPI approaches are designed to look at just that. They work by looking at the VOI of the remaining parameters of the model if we assume perfect information for the parameter of interest.
The EVPPI analyses often use a nested, double-loop Monte Carlo method, although alternative methods are able to approximate EVPPI using regression modelling and probabilistic analysis results [15, 16]. The double-loop method involves the parameter of interest being sampled in an outer loop, and all other parameters being sampled within the inner loop. For the case study, 100 inner and 100 outer loops are performed. For the EVPPI, each inner loop requires an estimation of the NMB of the two treatments, under the fixed parameter value of interest (i.e. the partial parameter).
In R, a similar function (named ‘nmb.function’) does this, by taking a vector of WTPs, and creates a matrix to estimate NMB, for each inner loop simulation, at each WTP. In contrast, in the MS Excel model, this is performed at just one WTP value, and uses the same structure as used to estimate the average NMB in the EVPI calculations. In both cases, the NMB for each treatment is stored from each inner loop.
4.5 Calculate EVPPI Values for WTP Values
In the ‘evppi.results.SP0’ and ‘evppi.results.NP1’ data frames in R, each row represents the mean NMB across all inner loop simulations. This is equivalent to cell AS6:AT105 in MS Excel, which is shown in Fig. 7 for comparison to the R code presented in the ESM. Each row represents the mean NMB (the average derived from all inner loop simulations), for each outer loop. In R, this is done across multiple columns, for each WTP. In MS Excel, this is done for just one WTP value.
Expected value of partially perfect information (EVPPI) calculations in Microsoft Excel (the equivalent sheet presenting formulas for each cell is presented in the ESM). OMRs rrNP1 = Relative risk of revision for new prosthesis 1 compared to standard OMRs = Operative mortality following primary and revision surgeries
cRevision = Costs (in this case the only probabilistic cost is cost of revision surgery)
Once all outer loop simulations have been performed, we can calculate the mean NMB across all outer loops simulations (‘current.info1’ and ‘current.info2’ in R, cells AS4:AT4 in MS Excel and Fig. 7). The treatment with the highest NMB is then chosen (‘current.info’ in R, in MS Excel, the equivalent is taken as part of the formula in cell AX7).
Next, using a similar approach taken to calculate EVPI, the highest NMB value across each simulation (i.e. each row) needs to be taken, to consider the decision made with perfect parameter information. In MS Excel, this is simply the higher value of the SP0 and NP1 columns (AS6:AT105, Fig. 7). The higher value is selected and shown in AU6:AU105 (Fig. 7). In R, the same calculation is done, but across multiple WTP values. First, an array is created, which in this case is essentially two data frames that contain the NMB from all outer loop simulations, across each WTP value. The apply function is then used to take the maximum NMB for either treatment (i.e. for any given simulation, at each WTP), to create a vector of maximum NMB values (‘perf.info.sims’).
Once the maximum NMB of either treatment is available for each iteration at each WTP, we can take the mean NMB across all simulations, for each WTP value (‘perf.info’ in R, AU4 in MS Excel). Finally, the EVPPI results are the difference between this partially perfect information, and current information, and then multiplied by the effective population. This is shown in AX7 in MS Excel (Fig. 7). In R, this is the data.frame ‘evppi.results’, which is returned from the ‘gen.evppi.function’. In this example, the R function returns the EVPPI at each of the WTP values, whereas in MS Excel, the value returned is for only one WTP value.
An example of how the EVPPI is calculated is shown in Table 3, for one partial parameter (re-revision risk) and at the £2500 WTP threshold only. Each row represents an outer loop. Once a re-revision risk value has been selected in the outer loop (in the first outer loop, it is 0.0414), then 100 inner loop simulations are performed with all other remaining parameters sampled across each inner loop. The mean NMB across inner loops, for the first outer loop, is estimated for each treatment, and reported. This is £36,287.04 for the standard prosthesis, and £36,305.70 for the new prosthesis.
The mean for each treatment is then estimated across all outer loops (£36290.53 for the new prosthesis and £36306.65 for the new prosthesis). The EVPPI for the re-revision risk is then estimated, taking the NMB under perfect information (£36306.83), and subtracting the highest NMB for either treatment (£36306.65), to give the difference between perfect parameter information and current information. This gives an EVPPI of £0.18, which multiplied by the effective population gives a population EVPPI of £55,985.
4.6 Run EVPPI Simulations
Now that we have functions to be able to process the results of the inner loops, and the outer loops to estimate the EVPPI, we can create the structure to run the EVPPI analyses. In R, we use three nested loops. The first loop (‘j’) selects the parameter of interest for the ‘partial’ perfect information. The number of ‘j’ loops are set to the number of partial parameters included in the EVPPI analyses. In this instance, we look at six parameter groups. Next, the outer loop (‘a’) will select a value for the partial parameter of interest, which will remain fixed within each inner loop. With each new outer loop, the parameter value for the partial parameter will change. The final loop is the inner loop (‘b’), in which all other model parameters are sampled (except the ‘partial’ parameter selected in the outer loop, which remains fixed).
Once the inner loop has completed, the mean NMB for each treatment will be estimated using the ‘nmb.function’ function. A new outer loop will proceed, in which a new parameter value for the partial parameter will be selected, and then the inner loops will be performed. Once all outer loops have been performed, the ‘gen.evppi.results’ function will estimate the EVPPI for this particular partial parameter. Finally, once the EVPPI for a parameter has been completed, a new partial parameter will be selected, and the process repeated, until all EVPPI results, for each partial parameter, have been performed. Note that in MS Excel, the selection of each outer loop partial parameter is performed in Visual Basic for Applications (see ESM).
The results of the EVPPI loops can be plotted. Note that in MS Excel, the EVPPI is only calculated at one WTP threshold at a time, and therefore, the values for each parameter are only shown at the particular WTP selected. Using R, we can plot the EVPPI over a range of WTP thresholds, see ESM for the code and plots related to graphical outputs.
5 Discussion and Conclusions
Taking the example of THR surgery from previously published MS Excel models, we provide a guide to building a Markov model in R that accounts for heterogeneity across population subgroups. We also provide a demonstration of how VOI analyses can be developed in R, with the R code compared directly to the corresponding Visual Basic for Applications code.
We also provide a demonstration of an economic model parameterised using a regression-based survival analysis. This allows the economic model to account for the heterogeneity that may exist in the survival analysis, creating samples based on covariates and covariance estimated through a parametric Weibull regression analysis. By building a flexible model that allows us to select the age and sex within the model function, we demonstrate how subgroup analyses can be easily performed, and integrated with such analyses. Survival analyses could easily precede the economic evaluation, with both being performed in R, allowing both the statistical and economic analyses to be performed using the same software. This avoids issues with reading in outputs externally, but also allows for all analyses and outputs to be updated and rerun simultaneously.
When comparing the time taken to run EVPPI loops, a total of 100 inner and 100 outer loops were performed for each of the six parameter groups included in the EVPPI analysis. This was approximately 11 times quicker in R than MS Excel, even when MS Excel calculated the EVPPI for only one WTP threshold, whereas this is calculated for 501 WTP values in R (£0–£50,000, in increments of £100). The speed advantage of R when running multiple simulations has already been noted [3]. Furthermore, alternative regression-based methods are available in R, which can be used to estimate EVPPI without the use of inner and outer loops (i.e. a double loop) [16]. The original MS Excel case study for which this R tutorial is based on does not utilise expected value of sample information techniques and therefore these analyses were not considered here. This is however an important tool in decision analyses, and other R guidance for such VOI analyses are available elsewhere [17, 18].
Other benefits of R over MS Excel highlighted by this direct comparison tutorial include: the ability to easily source other models and data, and the ability to readily conduct and store EVPPI results over multiple WTP thresholds. Additionally, by example, we highlight the ease of publishing and citing economic evaluation models in R open access through repositories such as Github [6]. Such models can also be adapted into Shinyapps and R packages [19, 20]. Whilst this tutorial utilises mainly base R, we introduce the concept of loading and using packages through the use of reshape and ggplot2 [9, 21]. A compilation of specific health economics packages can be found elsewhere [22].
In a world where the coronavirus disease 2019 pandemic and potential subsequent global recessions could lead to smaller healthcare budgets and funding available for research, VOI analyses within the healthcare technology appraisal process can provide a formal framework in quantifying the potential costs and benefits of gathering further information used in relevant health economic models. Additionally, ‘open source’ health economic modelling will increase general transparency and adaptability in the field [4].
We hope that this tutorial paper will help guide MS Excel users who may want, or need, to transfer their model to R. The publicly available code can provide a template for individuals to develop their own models that are able to capture heterogeneity amongst model subgroups, and are able to evaluate the VOI for a particular decision.
Notes
The outputted R list (‘sample.output’) equivalent to the parameter trial data in the “Simulation” sheet in the corresponding MS Excel model.
The <Hazard function> sheet within the downloadable MS Excel model equivalent utilises individual calculations to perform this process.
References
Incerti D, Thom H, Baio G, Jansen JP. R you still using excel? the advantages of modern software tools for health technology assessment. value Health. 2019;22(5):575–79.
Baio G, Heath A. When simple becomes complicated: why excel should lose its place at the top table. Glob. Reg Health Technol. Assess. 2017;4(1):grhta-5000247.
Hollman C, Paulden M, Pechlivanoglou P, McCabe C. A comparison of four software programs for implementing decision analytic cost-effectiveness models. PharmacoEconomics. 2017;35(8):817–30.
Nathan Green N, Lamrock F, Naylor NR, Williams J, Briggs A. Health economic evaluation using Markov models in R for microsoft excel users: a tutorial (2022, submitted).
Briggs A, Sculpher M, Dawson J, Fitzpatrick R, Murray D, Malchau H. The use of probabilistic decision models in technology assessment: the case of total hip replacement. Appl Health Econ Health Policy. 2004;3(2):79–89.
Excel-R-tutorials. Markov_Extensions. 2022. https://github.com/Excel-R-tutorials/Extensions_Paper. Accessed 20 Apr 2022.
Stephen N. Version control with Git and SVN. https://support.rstudio.com/hc/en-us/articles/200532077-Version-Control-with-Git-and-SVN. Accessed 20 Apr 2022.
Dragulescu AA. 2014. Read, write, format Excel 2007 (xlsx) files. https://mran.revolutionanalytics.com/snapshot/2015-07-13/web/packages/xlsx/vignettes/xlsx.pdf. Accessed 27/10/2022.
Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21(12):1–20.
Wickham H. Elegant graphics for data analysis. Media. 2009;35(211):10.1007.
Briggs A, Sculpher M, Claxton K. Decision modelling for health economic evaluation. 2022. https://www.herc.ox.ac.uk/downloads/decision-modelling-for-health-economic-evaluation. Accessed 20 Apr 2022.
Briggs A, Sculpher M, Claxton K. Decision modelling for health economic evaluation. Oxford: Oxford University Press; 2006.
R-Bloggers. Loading packages efficiently. 2019. https://www.r-bloggers.com/2019/10/loading-packages-efficiently/. Accessed 1 Nov 2021.
Alarid-Escudero F, Krijkamp EM, Enns, EA, Yang A, Hunink M, Pechlivanoglou P, Jalal H. An introductory tutorial on cohort state-transition models in R using a cost-effectiveness analysis example. 2020. https://www.arXiv preprint arXiv:2001.07824.
Rothery C, Strong M, Koffijberg HE, Basu A, Ghabri S, Knies S, Fenwick E. Value of information analytical methods: report 2 of the ISPOR value of information analysis emerging good practices task force. Value Health. 2020;23(3):277–86.
Strong M, Oakley JE, Brennan A. Estimating multiparameter partial expected value of perfect information from a probabilistic sensitivity analysis sample: a nonparametric regression approach. Med Decis Making. 2014;34(3):311–26.
Heath A, Kunst N, Jackson C, Strong M, Alarid-Escudero F, Goldhaber-Fiebert JD, Jalal H. Calculating the expected value of sample information in practice: considerations from 3 case Studies. Med Decis Making. (2020);40(3):314–26
Kunst N, Wilson ECF, Glynn D, Alarid-Escudero F, Baio G, Brennan A. Collaborative Network for Value of, I. Computing the expected value of sample information efficiently: practical guidance and recommendations for four model-Based methods. Value Health. (2020);23(6):734–42.
Smith R, Schneider P. Making health economic models Shiny: a tutorial. Wellcome Open Res. 2020;5:69.
Filipović-Pierucci A, Zarca K, Durand-Zalesk I. Markov models for health economic evaluations: the R package Heemod. arXiv preprint. 2017;1702.03252.
Wickham H. ggplot2: elegant graphics for data analysis Springer-Verlag New York; 2009. 2016
SHEPRD. Signposting Health Economic Packages in R for Decision Modelling (SHEPRD). 2021. https://hermes-sheprd.netlify.app/. Accessed 6 Oct 2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Financial support for this study was partially provided by an LSHTM seed funding grant. The views expressed are those of the author(s) and are not necessarily those of author-affiliated institutions, including the National Institute for Health Research, the UK Health Security Agency or the Department of Health and Social Care.
Conflicts of interest/Competing interests
Nichola R. Naylor, Jack Williams, Nathan Green, Felicity Lamrock and Andrew Briggs have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The data used for this tutorial paper are available at https://github.com/Excel-R-tutorials/Extensions_Paper.
Code availability
The code is available at https:// github. com/ Excel-R-tutor ials/ Exten sions_ Paper. as well as the data.
Authors’ contributions
JW: conceptualisation, writing original draft, validation, writing review and editing. NN: conceptualisation, writing original draft, validation, writing review and editing. NG: validation, writing review and editing. FL: validation, writing review and editing. AB: resources, writing review and editing.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Naylor, N.R., Williams, J., Green, N. et al. Extensions of Health Economic Evaluations in R for Microsoft Excel Users: A Tutorial for Incorporating Heterogeneity and Conducting Value of Information Analyses. PharmacoEconomics 41, 21–32 (2023). https://doi.org/10.1007/s40273-022-01203-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40273-022-01203-0