Abstract
Background and Objective
Several studies have demonstrated the ability to detect adverse events potentially related to multiple drug exposure via data mining. However, the number of putative associations produced by such computational approaches is typically large, making experimental validation difficult. We theorized that those potential associations for which there is evidence from multiple complementary sources are more likely to be true, and explored this idea using a published database of drug–drug-adverse event associations derived from electronic health records (EHRs).
Methods
We prioritized drug–drug-event associations derived from EHRs using four sources of information: (1) public databases, (2) sources of spontaneous reports, (3) literature, and (4) non-EHR drug–drug interaction (DDI) prediction methods. After pre-filtering the associations by removing those found in public databases, we devised a ranking for associations based on the support from the remaining sources, and evaluated the results of this rank-based prioritization.
Results
We collected information for 5983 putative EHR-derived drug–drug-event associations involving 345 drugs and ten adverse events from four data sources and four prediction methods. Only seven drug–drug-event associations (<0.5 %) had support from the majority of evidence sources, and about one third (1777) had support from at least one of the evidence sources.
Conclusions
Our proof-of-concept method for scoring putative drug–drug-event associations from EHRs offers a systematic and reproducible way of prioritizing associations for further study. Our findings also quantify the agreement (or lack thereof) among complementary sources of evidence for drug–drug-event associations and highlight the challenges of developing a robust approach for prioritizing signals of these associations.
Similar content being viewed by others
Prioritizing drug–drug-event association predictions for further evaluation is very important in pharmacovigilance because it is not feasible to experimentally validate very large numbers of predictions. |
We proposed a proof-of-concept approach to prioritize drug–drug-event associations derived from Electronic Health Records (EHRs) based on multiple sources of evidence. |
Our approach produced a ranked list of drug–drug-event associations for further investigation. |
1 Background and Significance
In the US, the annual cost of drug-related morbidity and mortality was estimated to be US$177.4 billion and rising in 2000 [1]. In 2012 alone (the most recent year for which this data is available from the Agency for Healthcare Research and Quality) there were more than 1.9 million emergency department visits in the US for adverse drug reactions [2]. Drug–drug interactions (DDIs) on a global scale are estimated to cause 0.57–4.8 % of all hospital admissions [3], and with around 70 % of interactions being clinically relevant and contributing to the majority of adverse drug reactions [4], there is a need for timely detection of potential DDIs that may cause adverse events (AEs). Adverse drug interactions occur when two or more drugs that are safe and effective when prescribed individually, pharmacokinetically or pharmacodynamically influence each other when taken concomitantly, resulting in reduced effectiveness, increased toxicity, or any other unintended harmful event. When new drugs are tested via in vivo and in vitro methods [5] before market approval, it is infeasible to test every possible interaction with all other existing and experimental drugs. An additional complicating factor is that some drugs require multiple periods of exposure for their adverse effects to become evident [6].
With the increasing use of electronic health records (EHRs) for research, researchers such as Wang et al. [7] have demonstrated the feasibility of using EHRs for pharmacovigilance. In related work, Harpaz et al. [8] used regression methods to detect adverse drug events from EHRs. Most previous EHR-based approaches have focused on finding associations between AEs and single drugs [9–12]. Prior to the widespread adoption of EHRs, researchers also used claims and billing data for drug safety surveillance [13, 14]. Related approaches (e.g., Shetty and Dalal [15]) use scientific literature repositories to detect signals of adverse drug events and validate their findings using EHRs (Duke et al. [16]).
Such computational approaches produce thousands of statistically plausible associations [17, 18], and experimentally validating all of them is not possible. In this work, we focused on drug safety surveillance using EHR data—particularly on associations between pairs of drugs and AEs. We proposed a proof-of-concept approach for systematically prioritizing putative drug–drug-event associations by mining multiple sources of evidence, filtering out known associations, and ranking these associations based on the number of sources that support a given association. We focused our study on 5983 putative EHR-derived drug–drug-event associations involving 345 drugs and ten adverse events previously reported by Iyer et al. [19]. Iyer and colleagues’ method is based on comparing the number of patients exposed to a pair of drugs and who experienced an AE to patients exposed to at most one drug (or neither of the two drugs) and experienced the event. In the original work, performance was evaluated on a gold standard of 1698 DDIs curated from DrugBank and Medi-Span, and found to have and area under the receiver operator characteristic (ROC) curve >80 %. The method was applied on two independent EHR datasets and found to perform similarly.
In this work, we distinguished drug–drug-event associations from DDIs. A drug–drug-event association refers to a medical manifestation that is associated with the concomitant use of two drugs but not necessarily causally related. For example, a variety of studies [20–24] have developed methods for detecting drug–drug-adverse event associations using spontaneous reporting systems (SRSs) such as the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS). We use the term DDI when there is evidence that two drugs are interacting with each other. There are methods that focus on identifying possible DDIs via molecular structure [25–28], interaction profiles [25, 29], drug target and side-effect similarities [27, 30], as well as similarity of known AEs based on drug interactions that result from a common metabolizing enzyme such as cytochrome P450 [17]. We used these methods as one source of evidence for drug–drug-event associations.
In order to rank EHR-derived drug–drug-event associations and prioritize their further investigation, we quantified support for a given association by first filtering out known associations using public databases and then using six sources of evidence divided into three categories: spontaneous reports, literature, and non-EHR-based prediction methods. The assumption behind our analysis is that putative drug–drug-event associations that are supported by multiple sources and methods are more likely to represent a true signal. We considered all drug–drug-event associations found in public databases at the time of our analysis to be ‘true’, and therefore removed them from the set of associations we prioritized. To obtain maximum coverage across the heterogeneous evidence sources, we developed dictionaries of drugs and AEs using identifiers and alternative names from multiple medical terminologies. We then implemented customized and previously developed methods for collecting evidence for drug–drug-event associations, using our dictionaries to relate the gathered evidence to the EHR-derived associations.
2 Materials and Methods
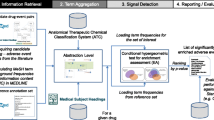
Our proof-of-concept approach for prioritizing drug–drug-event associations involved first collecting evidence of associations from a variety of drug–drug-event association and DDI prediction methods (or sources), filtering by removing DDIs found in public databases, and then aggregating this evidence to calculate a prioritization score for individual drug–drug-event associations. Our analysis was scoped to a set of 5983 putative drug–drug-event associations among 345 approved drugs and ten adverse events identified by Iyer et al. (described in the Sect. 3) because our primary aim was to devise an approach to prioritize associations found from EHRs. In the following sections, we first summarize the method used by Iyer et al. [19] to identify potential drug–drug-event associations using EHR data. We then describe each of the prediction methods we used to collect evidence for each candidate drug–drug-event association, including details of our implementation of methods where they differed from previously published work. Lastly, we describe the results of this process and how we aggregated the data to generate a prioritization score for each drug–drug-event association examined. Figure 1 provides an overview of our approach, illustrating the four categories of sources and methods we used for prioritization.
Overview of sources used for prioritizing drug–drug-event associations. EHR-derived associations are used as input to search existing evidence sources (1–3), and to assess support from previously developed DDI prediction methods (4). We group the information and DDI prediction methods into 1 public databases (green)—used for filtering out known associations; 2 official sources of drug adverse event reports (yellow); 3 biomedical literature (pink); and lastly 4 non-EHR-based DDI prediction methods. For each evidence type, we also show the specific sources we used and methods implemented, respectively. Only one of the four DDI prediction methods (TWOSIDES) associates predicted interactions with ADEs (cyan). The other three methods predict drug–drug interactions without an accompanying ADE (orange). ADE adverse drug event, DDI drug–drug interaction, EHR electronic health records
While our current methodology does not distinguish between missing data in a given source and evidence of no association, as an inclusion criterion during the curation step of the data sources we ensured that at least 80 % of the unique drugs found in the EHR-derived list of drug–drug-event associations were present in each source. Our assumption was that if the drug is present in a source, any associations it participates in will also be covered.
2.1 Drug–Drug-Event Associations Derived from EHR Data
In previous work, Iyer et al. devised a method for identifying candidate drug–drug-event associations from EHR data by constructing a contingency table as typically used in drug safety surveillance studies. The number of patients exposed to a pair of drugs and experiencing an AE was compared with that of patients exposed to at most one drug (or neither of the two drugs) that experienced the event or did not experience the event (Fig. A in the electronic supplementary material). The assignment of a patient to one of the four cells in the contingency table was based on occurrence and temporal order of the first mention of the drugs and AE in the patient’s record, ignoring drug mentions that appeared after the first occurrence of the event. A raw association score for exposure to the two drugs and occurrence of the event was calculated, in the form of an odds ratio, based on the patient counts in the contingency tables. An adjusted odds ratio (AOR) was calculated for each drug–drug-event association after matching exposed and comparison group patients on age, gender, number of co-morbidities, and number of unique drugs in their record, and then performing a conditional logistic regression. The lower bound of the 95 % confidence interval of the AOR was used as the final association score. An association was determined to be significant if the final score was greater than a threshold value yielding the desired sensitivity and specificity, based on a ROC curve constructed from a gold-standard set of known true positive DDIs and negative non-interactions. The 5983 drug–drug-event associations reported in the work were limited to associations involving drugs prescribed to a minimum of 100 patients in the EHR data. Individual prevalence details for these associations and a comprehensive description of the method used for predicting drug–drug-event associations from EHR data can be found in [19]. In this work, we propose a proof-of concept method to further prioritize these associations by filtering out known associations and aggregating evidence from complementary sources of evidence in order to identify a subset of associations as candidates for further investigation.
2.2 Building a Dictionary of Drugs and Adverse Events
To enable a systematic search of complementary evidence sources for drug–drug-event associations, we built a dictionary of names and identifiers for the 345 drugs and ten AEs in our study. Our goal was to identify all possible identifiers and alternative names for each drug and AE from multiple terminologies.
For drugs, we first used string matching to map the drug names provided by Iyer et al. to RxNorm and DrugBank drug name identifiers. We then used the RxNorm identifiers to derive additional mappings to UMLS (Unified Medical Language System) Concept Unique Identifiers (CUIs), SNOMED (Systematized Nomenclature of Medicine) Clinical Terms (CT) concepts, MeSH codes, and DrugBank identifiers. Lastly, we integrated all identifiers and alternative names into one drug dictionary, to enable the mapping between all sources covered.
We used a similar approach for the ten AEs in our study: we first mapped each AE by string matching to UMLS CUIs, and from these CUIs to SNOMED CT concepts and MeSH codes. From these sources, we collected all identifiers and alternative names (including synonyms) and compiled them into one AE dictionary.
We used the terms and identifiers in our dictionaries to match drugs and AEs for our study in the sources and prediction method results described in the next section.
2.3 Sources and Methods Used for Prioritizing Associations Derived from EHRs
2.3.1 Public Databases
The associations found in these public databases were used to filter the associations derived from EHRs as they are likely to be true since they appear in professionally curated resources that report known drug–drug associations with given indications. In total, we removed 201 associations from our set of 5983 DDIs, leaving us with 5782 DDIs for further study.
2.3.1.1 DrugBank
Using the same approach as Iyer et al. [19], we extracted known drug–drug-event associations from DrugBank [31]. We used a Linked Data version of DrugBank made publicly available through the Bio2RDF open-source project [32], dated 25 July 2013. Drug pairs were extracted from the structured data describing DDIs in Bio2RDF’s DrugBank dataset, and drug names were directly matched to our dictionary using the UMLS ‘preferred term’ to resolve ambiguity in the case that more than one drug from our dictionary was matched. The AEs were identified by applying the text annotation pipeline used in Iyer et al. [19] to the manually curated description of the given DDI and the resulting UMLS CUIs were matched to our event dictionary.
2.3.1.2 Drugs.com
We used web crawling agents to process Drugs.com for its DDI data. We programmatically searched all Drugs.com pages for any of the drug names from our drug dictionary and automatically extracted all reported DDIs for any matched drug, following the hyperlink to the drug’s ‘interactions’ website. From the resulting list of interactions for each drug, we identified those that matched any of the drug–drug pairs from the 5983 associations. We then processed for each interaction the information sections for consumers and professionals using the text annotation pipeline as described above to identify AEs, and then mapped them to our dictionary via the UMLS CUIs produced by the text annotation pipeline. Note that we did not take into account the severity of the AE in this process. To ensure that we did not incorrectly extract drug–drug-event associations due to adverse events reported only for single drugs, we only processed the text from the interaction section of a given drug’s page.
2.3.2 Sources of Reported Information
2.3.2.1 FDA Adverse Event Reporting System (FAERS)
We analyzed over 3.2 million reports from FAERS from 2004 to 2014 in order to find support for drug–drug-event associations. We analyzed the data by counting reports that matched a given pair of drugs and the AE specified in a putative association, disregarding the reporting source. Event names from FAERS were mapped to our AE dictionary via their Medical Dictionary for Regulatory Activities (MedDRA®) label, and drug names were matched directly to names in our drug dictionary. We combined the drug and reaction tables from FAERS by linking drugs and events via their case identifiers, and identified all resulting associations. We required at least ten unique FAERS reports to signal support for a given drug–drug-event association. We empirically selected ten as the minimum number of reports required because drug–drug-event associations from the gold standard of known drug–drug-event associations from Iyer et al. [19] had on average ten reports in FAERS. We use this conservatively high threshold to take into account that a report in FAERS may list co-administered drugs in addition to any drugs suspected to be responsible for an observed adverse event, which as a result can be incorrectly identified as co-suspect drugs. We do not explicitly distinguish between co-administered and co-suspect drugs, but given the threshold of ten reports, only those drug pairs that appear consistently in reports with a specific event will be detected using this method.
2.3.3 Literature
2.3.3.1 MEDLINE®
To mine MEDLINE for evidence supporting drug–drug-event associations, we extended a method proposed in Avillach et al. [33] that uses MeSH annotations of MEDLINE articles for detecting relationships between individual drugs and AEs. Our approach consisted of two steps: (1) retrieve relevant articles from MEDLINE using a customized search query and (2) extract the drug–drug-event sets from the resulting articles based on their MeSH index terms. We retrieved 10,459 articles (as of 28 April 2015) using the PubMed Entrez Programming utilities with the following query (our modification of the original query is highlighted in bold):
“Chemicals and Drugs Category/adverse effects”[Mesh] AND “chemically induced”[Subheading] AND “Drug interactions”[Mesh].
For this study we considered articles indexed with qualified MeSH terms corresponding to the 365 drugs and ten events of interest through our drug and AE dictionary. For example, we found evidence for the drug–drug-event association linezolid–venlafaxine–serotonin syndrome based on five articles with the following MeSH term/subheading combinations: the drugs linezolid (mapped to oxazolidinones/adverse effects) and venlafaxine (with pharmacological action serotonin uptake inhibitors/adverse effects), and the event serotonin syndrome (mapped to serotonin syndrome/chemically induced).
2.3.4 Non-EHR-Based Drug–Drug-Interaction Prediction Methods
We also used four previously developed DDI prediction methods to identify supporting evidence for the set of 5983 EHR-derived drug–drug-event associations. One of these methods (TWOSIDES) predicts DDIs and associations with AEs for these interactions, while the other three (INDI [INferring Drug Interactions], the similarity-based modeling method, and the predictive pharmacointeraction networks approach) predicts DDIs only (e.g., physical interaction or changes in the rates of drug metabolism) and do not predict specific AEs. Practically, we counted support from methods of the latter type for each EHR-derived drug–drug-event association in which the pair of drugs match, independent of the corresponding AE. We describe each of the four prediction methods in more detail in the following sections.
2.3.4.1 TWOSIDES
TWOSIDES [18] is a database that contains predicted side effects associated with pairs of drugs. It provides 4,651,131 potential drug–drug-event associations mined from FAERS in the period of the first quarter of 2004 through to the first quarter of 2009. The drug–drug side effects in this database are a subset of FAERS-reported associations, filtered using a method aimed to control for unmeasured confounding factors and reporting bias in a data-driven manner. We matched drug names and AE UMLS CUIs from TWOSIDES to our dictionary.
2.3.4.2 INferring Drug Interactions (INDI)
INDI [17] is a DDI prediction method that can infer both pharmacokinetic and pharmacodynamic interactions based on similarity to known interactions that result from a common metabolizing enzyme (CYP). The original set of predictions contains 57,746 drug–drug pairs, of which 4185 involve the 345 drugs from the set of EHR-derived drug–drug-event associations. Our method signaled a ‘match’ if both the drugs in the predicted interaction matched the drugs from an EHR-derived drug–drug-event association. For example, we searched INDI predictions for the EHR-derived association rosuvastatin–caspofungin–rhabdomyolysis by only looking for terms from our drug dictionary for rosuvastatin–caspofungin. We match drugs from INDI to our dictionary via UMLS CUIs.
2.3.4.3 Similarity-Based Modeling
A method recently published by Vilar et al. [28] uses a gold standard of DDIs and drug similarity information extracted from multiple sources—including 2D and 3D molecular structure, drug target similarities, and side-effect similarities—to learn salient characteristics of interacting drugs. One advantage of this method is that it is interpretable because it generates drug interaction candidates that are traceable to pharmacological or clinical effects. For this method, we collaborated with the authors to re-calibrate their method by providing them the gold standard used in Iyer et al. [19] and applying their method to the set of drugs from the EHR-derived associations. After this similarity recalibration (which will vary depending on the gold standard used), we determined the correct prediction thresholds (corresponding to 90 % specificity or greater) for selecting predictions with sufficient support.
2.3.4.4 Predictive Pharmacointeraction Networks
Developed by Cami et al. [34], this method predicts DDIs based on the structure of a network of drugs constructed from a knowledgebase of known DDIs. Using a network that consists of drugs as nodes and known interactions between these drugs as edges, a predictive model identifies additional edges that may represent novel DDIs. Collaborating with the authors, we matched 306 of the 345 drugs from our dictionary to the 856 drugs in their dataset, allowing a total of 4364 EHR-derived drug–drug-event associations to be analyzed. We excluded 1214 of these associations from analysis because they were previously reported as interactions in Multum VantageRx, a commercially available database of curated DDIs and side effect information used as the learning source by the authors of this method.
2.4 Drug Class Distribution of Analyzed Drugs
The 345 drugs in our study were mapped to RxNorm and UMLS CUIs, as previously described. RxNorm links drug ingredients to the Anatomical Therapeutic Chemical (ATC) classification [35]. ATC is a system developed by the World Health Organization (WHO) Collaborating Centre for Drug Statistics Methodology that classifies drugs at five levels: anatomical (1st), therapeutic (2nd), pharmacological (3rd), chemical (4th), and ingredient (5th). In order to establish drug class membership for the 345 drugs in this study, we mapped these drugs to ingredients at the 5th level in ATC through RxNorm and then aggregated them into drug classes according to the hierarchical information in ATC. We aggregated the 345 drugs into 67 2nd level classes (therapeutic main group) and finally into 14 1st level classes (anatomical main group) according to the ATC code hierarchy.
The 345 drugs in the drug–drug-event associations are distributed across both anatomical and therapeutic classes (Fig. 2). A high proportion of the drugs can be grouped into three anatomical classes: drugs acting in the nervous system [67], in the cardiovascular system [67], and antineoplastic and immunomodulating agents [66]. In terms of their therapeutic intent, 49 drugs are classified as antineoplastic agents (ATC L01).
Distribution of ATC classes for the 345 drugs for which drug–drug-event associations were derived from the electronic health record (EHR) by Iyer et al. [19]. The horizontal axis shows the number of drugs classified in each anatomical (1st level) and therapeutic (2nd level) class (multiple classifications are possible). Of the 345 drugs, 67 belong to the nervous system (N) and another 67 to the cardiovascular system (C) class. At the therapeutic level, the class with most drugs in the set (49) is antineoplastic agents (L01, in light blue)
2.5 Distribution of Adverse Events Across Drug–Drug-Event Associations
The 5983 drug–drug-event associations analyzed in this work are distributed across ten AEs: bradycardia, cardiac arrhythmia, hyperkalemia, hypoglycemia, long QT syndrome, neutropenia, pancytopenia, Parkinsonian symptoms, rhabdomyolysis, and serotonin syndrome (Table 1). These ten events are a subset of the proposed adverse events by the Exploring and Understanding Adverse Drug Reactions (EU-ADR) project, which had enough data support at Stanford.
2.6 Prioritization and Scoring of Drug–Drug-Event Associations
In order to provide a proof-of-concept prioritization of the EHR-derived drug–drug-event associations and to quantify the degree of supporting evidence for each association, we cross-referenced each association with the data sources and prediction methods described above. We first removed all associations found in public databases (Drugs.com and DrugBank), since they represent comprehensive repositories of known drug information, and then used the other sources to quantify supporting evidence for the remaining associations. We grouped sources providing similar information into categories of evidence, counting support from each source category rather than from each source individually, to avoid artificial inflation of scores. For each source category, if support was signaled we added a value of 1 to the score. Specifically, sources of spontaneous reports and literature each contributed 1 to the total score. We keep separate scores contributed from the prediction methods to distinguish predictions that specify an AE from predictions that specify only a DDI without a corresponding AE. Thus, for each drug–drug-event association the maximum score possible is 4. The four categories we used are:
-
(a)
Spontaneous reporting: If ten or more FAERS reports match a given drug–drug-event association then support is signaled for this association.
-
(b)
Literature: Using our adapted MEDLINE extraction method, we consider a drug–drug-event association to be supported if there are two or more articles whose MeSH index terms provide evidence for the association.
-
(c)
DDI prediction methods where the AE is specified (DDI + AE): In this category we include the methods that produce drug–drug-event predictions and signal support if both drugs and the AE for a drug–drug-event association match a given prediction.
-
(d)
DDI prediction methods where the AE is not specified (DDI): In this category, we signal support for a drug–drug-event association if both drugs in the association match any of the predictions from the three methods implemented.
We ranked the associations by their total score to identify those associations with the highest degree of support from complementary sources.
3 Results
Of the 5983 analyzed associations, 201 associations were described in public databases, so we removed them from our list to prioritize. Overall, only a small subset of the associations had support from the majority of evidence sources. However, there is a positive correlation between the number of sources providing support for EHR-derived associations and the association scores as calculated by Iyer et al. [19]. Interestingly, we also found that there is low agreement among the evidence sources supporting a given association, which highlights the challenge of developing comprehensive methods for predicting DDIs and drug–drug-event associations. Such low agreement among sources was also recently noted by Ayvaz et al. [36], who created a dataset of known DDIs from publicly available sources.
3.1 Distribution of Prioritization Scores and Sources of Support Across Drug–Drug-Event Associations
Figure 3 shows the distribution of prioritization scores and contributions from each evidence source for all drug–drug-event associations, as well as the median adjusted odds ratio from Iyer et al. for the drug–drug-event associations with a given prioritization score. Non-EHR-based prediction methods (for both DDIs and drug–drug-event associations) contributed to a high proportion of the support for drug–drug-event associations across all prioritization score values (blue outline box), while reporting sources lent support to all (7 out of 7) drug–drug-event associations with a score of 3 or 4 (green outline box). Reporting sources (e.g., FAERS) contributed support to a small proportion of the drug–drug-event associations overall—this may be due to our conservative threshold of requiring ten reports in FAERS for the drug–drug-event association to be considered. This could be adjusted using a similar analysis with a different gold standard. Non-EHR-based prediction methods provided evidence for the greatest number of drug–drug-event associations, while literature provided evidence for the fewest.
Contribution of evidence sources across prioritization scores. The number in each square is the number of drug–drug-event associations with a given score (shown on the left vertical axis) that had support from a particular source (shown on the bottom horizontal axis). The last column shows the median adjusted odds ratio for drug–drug-event associations in the given row. Squares with greater red intensity indicate that a high proportion of drug–drug-event associations with that score value (row) had support from that source (column). Non-EHR-based prediction methods supported a high proportion of the drug–drug-event associations across all prioritization score values (blue outline box), while spontaneous reporting lent support to all (6 out of 6, green outline box) drug–drug-event associations with a score of 3 or 4. AE adverse event, DDI drug–drug interaction, EHR electronic health records
Analyzing the contribution of evidence sources to drug–drug-event associations with a given prioritization score offers a means to select associations for further investigation. For example, all seven drug–drug-event associations with a prioritization score ≥3 had support from reporting sources (FAERS). Adjusted odds ratio (AOR) for drug–drug-event associations as computed by Iyer et al. [19] also correlate with the prioritization score. The median AOR for drug–drug-event associations increases as prioritization score increases (Fig. 3).
The highest scoring drug–drug-event associations (with a score ≥3, indicating support from a majority of sources) are listed in Table 2.
3.2 Distribution of Support for Adverse Events
Figure 4 shows the distribution of AEs across the data sources we use to prioritize drug–drug-event associations. Neutropenia is the most frequently occurring AE across all associations, and long QT syndrome is the least frequent. Interestingly, among all 5983 predicted drug–drug-event associations, only those drug interactions associated with hypoglycemia are described in public databases (green) and therefore filtered out.
3.3 Low Consensus of Support Across Sources
In order to examine the consensus across multiple sources and prediction methods we quantified the overlap of drug–drug-event associations or DDI predictions among them. We restricted the drug and event space to the 345 drugs and ten AEs of interest in this study in order to provide a comprehensive comparison in the context of prioritizing EHR-derived associations and to avoid having lower overlap ratios purely as a result of analyzing drug and event associations with source-dependent differences.
Table 3 shows that the sources and prediction methods that provided supporting evidence for prioritizing drug–drug-event associations are highly variable in the degree of overlap of their content. For some sources, the degree of overlap is very low. For example, for DrugBank and MEDLINE only one association is supported by both sources. Such low agreement among sources was also recently noted by Ayvaz et al. [36], while creating a dataset of known DDIs from public sources.
4 Discussion
4.1 Challenges in Prioritization
The first hurdle in terms of performing a source and method-wide evidence gathering and prioritization approach, such as the one we present here, lies in the heterogeneity of the resources and methods. A great deal of effort was required to normalize drug and AE names and identifiers, as well as to subset the data to a manageable drug universe and obtain comparable outputs for analysis. The second challenge was devising a scoring scheme that groups the evidence sources in a balanced way and provides good coverage across drug classes, event types, and evidence sources. As a research task, prioritizing drug–drug-event associations is complicated by the diversity of the methods used to generate possible drug–drug-event associations, as well as their differing coverage of the drug and event spaces.
Our work addresses these challenges and enables the prioritization of drug–drug-event associations from a given source. The prioritization scheme can also be easily modified and improved to use alternative groupings or weighting of contributions from the different methods and sources used. We use a simple and transparent grouping of similar types of sources and a direct sum of their contributions as a score for each drug–drug-event association. We do not differentially weight the contribution of any one evidence source to favor its underlying mechanism, in this first foray into developing a comprehensive prioritization method.
One possible way to enhance our prioritization scheme is to devise a more finely tuned method for mining the literature for drug–drug-event associations. Our current approach does not detect associations that are not captured in MeSH annotations. Our method infers associations from the co-occurrence of index terms—some of which may not have been intended by MeSH indexers—and therefore may produce false positive associations. Advanced text mining on article abstracts or full texts may be helpful in distinguishing drug–drug-event associations from drugs individually reported to be related to an event, for instance in comparative studies.
4.2 Manual Review of the Proof-of-Concept Prioritization
Among the seven drug–drug-event associations with a score >3 (in Table 2), three associations involve neutropenia. The first two, cyclophosphamide–carboplatin–neutropenia and cyclophosphamide–cisplatin–neutropenia, have support from FAERS reports, literature, and non-EHR-based prediction methods. On a manual review of the supporting evidence, the literature-based evidence is the weakest. In the two articles whose MeSH indexing lent support for these associations, neutropenia is associated with chemotherapy (and thus indexed with the MeSH subheading ‘chemically induced’), but the adverse events discussed in these articles are nausea and vomiting. For association #3, thalidomide–warfarin–neutropenia, we find that thalidomide and warfarin are known to interact (as reported in public databases—Drugs.com), but the event is not listed. Based on the EHR-derived associations and the prediction methods, we now have a possible event with which the two drugs could be associated. Association #4, simvastatin–gemfibrozil–rhabdomyolysis, is the subject of two correctly indexed MEDLINE articles. Thus, we regard the support from the literature as correct. Association #5, digoxin–carvedilol–hyperkalemia, is found in at least one public database (Drugs.com), without the matching adverse event. Support from other sources (reporting and prediction methods) indicates that hyperkalemia might be a possible adverse event for these two drugs. Association #6, spironolactone–atenolol–hyperkalemia, has some support from online sources [37] and [38], but is not found in DrugBank or Drugs.com. Lastly, for association #7, spironolactone–glimepiride–hyperkalemia, there are no mentions of this association in the literature or public databases, but an online source [39] offers support for this association.
4.3 Alternative Groupings for Prioritization
We also devised two alternative ways of grouping our data sources to produce prioritization rankings (see electronic supplementary material, Tables I and II). For the first alternative grouping, we removed all grouping constraints, counted each source and prediction method separately, and assigned equal weight to each. With a maximum possible score of 6, we found one potential association with a score of 5. This association is the same as the top ranked association presented in Table 3. All other associations in electronic supplementary material Table I have a score between 4 and 5, for a total of 22—a significant increase from the 7 top scoring associations in Table 3. The stability of the ranking is evidenced by the fact that all the associations found in Table 3 are found in the same relative order (though not rank position) in electronic supplementary material Table I. The effect of equally weighting all sources is that more associations surface as being higher ranked.
For our second alternative grouping we combined the support from the prediction method TWOSIDES with spontaneous reporting sources as they both rely on the FAERS as their source of information (electronic supplementary material, Table II). We considered all prediction methods as one group and considered literature (MEDLINE) as one separate group. With such a grouping, the maximum score possible is 3, and two associations received this score. These two associations are the top two ranked associations from Table 3 (the original grouping) and associations #1 and #3 from electronic supplementary material Table I. This again demonstrates the stability of the highest scored associations. This three bucket grouping resulted in over 187 associations having a score of 2, making the task of ranking them within each score group harder.
A number of alternative groupings and differing weight assignments can be used with our approach. However, in the three scenarios we have considered, we find that highly ranked associations stay that way, suggesting that these associations warrant further investigation.
An interesting alternative for grouping and ranking would be to prioritize based on the severity of adverse events, and identify which associations contain the most severe adverse events. However, given the rarity of severe adverse events, the challenges in unambiguous attribution of adverse events to a given set of drugs, and lack of a common ‘grading scheme’ for severity, we avoid prioritizing based on severity in this work.
4.4 Limitations
Our current approach is limited by the information provided by non-EHR-based methods and sources available and their own unique approach for detecting signals of drug–drug-event associations or making predictions. Their underlying design may not be ideal for some of the EHR-derived drug–drug-event associations we attempt to prioritize. Such a mismatch can result in smaller scores for some associations because their corresponding drugs and events are underrepresented in the sources or prediction methods we have incorporated into our proof-of-concept prioritization scheme.
The scoring scheme we propose is also dependent on the types of evidence sources available. For example, to incorporate information from methods that predict DDIs with a specific AE as well as those that predict only DDIs, we separated these methods into two sub-categories that contributed scores independently. We believe that by further incorporating more diverse sources and methods we can reduce the coverage gap of specific AEs at the expense of having more categories of evidence.
It should also be noted that information derived from different data source categories might be correlated. For instance, public databases may have entries about drug interactions that are based on published information from the literature. However, in our study we did not detect such a systematic overlap between supporting information from different categories. On the contrary, one of our discoveries in this study is that there is little overlap between different data sources and methods—thus underscoring the need to examine multiple source types when prioritizing a set of drug–drug-event associations for plausibility and further investigation.
In this study, we restrict drug–drug associations to pairs of drugs. However, it is known that there are cases where AEs are associated with the concomitant use of more than two drugs. For instance, Maxa et al. report a case of rhabdomyolysis after concomitant use of cyclosporine, simvastatin, gemfibrozil, and itraconazole [40]. In this case, it would be misleading to report the event, rhabdomyolysis, individually for all pairs of two drugs from these four (e.g. cyclosporine–simvastatin–rhabdomyolysis), since a given pair may be necessary but not sufficient to cause the effect. AEs associated with many drugs taken at the same time are common in cancer treatment (antineoplastic agents). Such multi-drug adverse events are not covered in our current analysis.
Lastly, using MeSH indexing can introduce false positive evidence for a given association. As described in Sect. 4.2 for associations #1 and #2, an inspection of the articles supporting these associations revealed the potential for ambiguity. The MeSH index terms for these articles listed more than a single pair of drugs with the adverse effects subheading. Thus, literature support for prediction #1 and #2 should be considered false positives resulting from the cross-product of all disease terms with a ‘chemically induced’ qualifier and drugs with adverse effects in a given article. We believe that advanced text mining on article abstracts or full text may be helpful in distinguishing drug–drug-event associations from drugs individually reported to be related to an event, and also interacting with another drug in the context of a different adverse event.
4.5 Related Work
Recently, there have been a number of community efforts to combine publicly available data sources of drug–drug-event associations [36], rank adverse drug reactions [41], and to manually evaluate DDI evidence for inclusion in Clinical Decision Support tools [42]. There are also working groups within the Observational Health Data Sciences and Informatics (OHDSI) collaborative that provide open-source standardized knowledge bases of the effects of medical products [43]. Our efforts provide a complementary source of prioritized, EHR-derived drug–drug-event associations and a framework for prioritizing drug–drug-event associations that is reproducible and easily automatable.
5 Conclusions
We have developed a proof-of-concept approach for scoring, and prioritizing putative drug–drug-event associations based on the degree of cmplementary evidence across multiple sources. We demonstrate that the ranking is stable under alternative grouping and weighting schemes. Given the large number of associations identified by individual approaches that signal DDIs and drug–drug-event associations, our work offers a systematic and reproducible way of prioritizing these associations for further study. Our findings also quantify the agreement (or lack thereof) among complementary sources of evidence for drug–drug-event associations and highlight the challenges in developing a robust approach for prioritizing signals of adverse drug events.
References
Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash, DC 1996). 2001;41(2):192–9.
Lucado J PK, Elixhauser A. HCUP National Emergency Department Sample (NEDS) - HCUP Statistical Brief #109. Agency for Healthcare Research and Quality. 2012. http://psnet.ahrq.gov/resource.aspx?resourceID=21786
Becker ML, Kallewaard M, Caspers PWJ, Visser LE, Leufkens HGM, Stricker BH. Hospitalisations and emergency department visits due to drug–drug interactions: a literature review. Pharmacoepidemiol Drug Saf. 2007;16(6):641–51.
Kiran P. Typical drug interactions in oncology. US Pharm. 2011;36(1):7–9.
Zhang L, Zhang Y, Zhao P, Huang S-M. Predicting drug–drug interactions: an FDA perspective. AAPS J. 2009;11(2):300–6.
Triaridis S, Tsiropoulos G, Rachovitsas D, Psillas G, Vital V. Spontaneous haematoma of the pharynx due to a rare drug interaction. Hippokratia. 2009;13(3):175–7.
Wang X, Hripcsak G, Markatou M, Friedman C. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc JAMIA. 2009;16(3):328–37.
Harpaz R, Perez H, Chase HS, Rabadan R, Hripcsak G, Friedman C. Biclustering of adverse drug events in FDA’s spontaneous reporting system. Clin Pharmacol Ther. 2011;89(2):243–50.
Haerian K, Varn D, Vaidya S, Ena L, Chase HS, Friedman C. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther. 2012;92(2):228–34.
LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther. 2013;93(6):547–55.
Lependu P, Iyer SV, Fairon C, Shah NH. Annotation analysis for testing drug safety signals using unstructured clinical notes. J Biomed Semant. 2012;3(Suppl 1):S5.
Liu M, McPeek Hinz ER, Matheny ME, Denny JC, Schildcrout JS, Miller RA, et al. Comparative analysis of pharmacovigilance methods in the detection of adverse drug reactions using electronic medical records. J Am Med Inform Assoc. 2013; 20(3):420–6. doi:10.1136/amiajnl-2012-001119.
Dore DD, Seeger JD, Arnold Chan K. Use of a claims-based active drug safety surveillance system to assess the risk of acute pancreatitis with exenatide or sitagliptin compared to metformin or glyburide. Curr Med Res Opin. 2009;25(4):1019–27.
Nadkarni PM. Drug safety surveillance using de-identified EMR and claims data: issues and challenges. J Am Med Inform Assoc JAMIA. 2010;17(6):671–4.
Shetty KD, Dalal S. Using information mining of the medical literature to improve drug safety. J Am Med Inform Assoc. 2011; 18(5):668–674. doi:10.1136/amiajnl-2011-000096.
Duke JD, Han X, Wang Z, Subhadarshini A, Karnik SD, Li X, et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput Biol. 2012;8(8):e1002614.
Gottlieb A, Stein GY, Oron Y, Ruppin E, Sharan R. INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol. 2012;8:592.
Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31-ra31.
Iyer SV, Harpaz R, LePendu P, Bauer-Mehren A, Shah NH. Mining clinical text for signals of adverse drug–drug interactions. J Am Med Inform Assoc. 2014;21(2):353–62.
Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinform. 2010;11(Suppl 9):S7-S.
Kadoyama K, Miki I, Tamura T, Brown JB, Sakaeda T, Okuno Y. Adverse event profiles of 5-fluorouracil and capecitabine: data mining of the public version of the FDA adverse event reporting system, aers, and reproducibility of clinical observations. Int J Med Sci. 2012;9(1):33–9.
Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug–drug interactions in adverse event reports. J Am Med Inform Assoc JAMIA. 2012;19(1):79–85.
Thakrar BT, Grundschober SB, Doessegger L. Detecting signals of drug–drug interactions in a spontaneous reports database. Br J Clin Pharmacol. 2007;64(4):489–95.
van Puijenbroek EP, Egberts AC, Heerdink ER, Leufkens HG. Detecting drug–drug interactions using a database for spontaneous adverse drug reactions: an example with diuretics and non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 2000;56(9–10):733–8.
Bauer-Mehren A, van Mullingen EM, Avillach P, Carrascosa MdC, Garcia-Serna R, Piñero J, et al. Automatic filtering and substantiation of drug safety signals. PLoS Comput Biol. 2012;8(4):e1002457.
Vilar S, Harpaz R, Uriarte E, Santana L, Rabadan R, Friedman C. Drug–drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc. 2012;19(6):1066–74.
Vilar S, Uriarte E, Santana L, Tatonetti NP, Friedman C. Detection of drug–drug interactions by modeling interaction profile fingerprints. PLoS One. 2013;8(3):e58321.
Vilar S, Uriarte E, Santana L, Lorberbaum T, Hripcsak G, Friedman C, et al. Similarity-based modeling in large-scale prediction of drug–drug interactions. Nat Protoc. 2014;9(9):2147–63.
Vilar S, Harpaz R, Santana L, Uriarte E, Friedman C. Enhancing adverse drug event detection in electronic health records using molecular structure similarity: application to pancreatitis. PLoS One. 2012;7(7):e41471.
Liu M, Wu Y, Chen Y, Sun J, Zhao Z, X-w Chen, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc JAMIA. 2012;19(e1):e28–35.
Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(Database issue):D1091-D7.
Callahan A, Cruz-Toledo J, Ansell P, Dumontier M. Bio2RDF release 2: improved coverage, interoperability and provenance of life science linked data. In: Cimiano P, Corcho O, Presutti V, Hollink L, Rudolph S, editors. The semantic web: semantics and big data. lecture notes in computer science. 7882: Springer Berlin Heidelberg; 2013. p. 200–12.
Avillach P, Dufour J-C, Diallo G, Salvo F, Joubert M, Thiessard F, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU–ADR project. J Am Med Inform Assoc JAMIA. 2013;20(3):446–52.
Cami A, Manzi S, Arnold A, Reis BY. pharmacointeraction network models predict unknown drug–drug interactions. PLoS One. 2013;8(4):e61468.
Organization WH. Anatomical Therapeutic Chemical (ATC) classification system [cited 2015 May 1st]. http://www.whocc.no/atc/structure_and_principles/. Accessed 1 May 2015.
Ayvaz S, Horn J, Hassanzadeh O, Zhu Q, Stan J, Tatonetti NP, et al. Toward a complete dataset of drug–drug interaction information from publicly available sources. J Biomed Inform. 2015;55:206–17.
Index R-TID. Drug interactions with atenolol oral and spironolactone oral [cited 2015 August 10th]. http://www.rxlist.com/drug-interactions/atenolol-oral-and-spironolactone-oral-interaction.htm. Accessed 10 Aug 2015.
Information e-Ph. Review: taking Spironolactone and Atenolol together [cited 2015 August 10th]. Available from: http://www.ehealthme.com/drug-interactions/atenolol-and-spironolactone. Accessed 10 Aug 2015.
Information e-Ph. From FDA reports: drug interactions between Glimepiride, Spironolactone for a female patient aged 73 [cited 2015 August 10th]. http://www.ehealthme.com/Spironolactone-Glimepiride-3847893. Accessed 10 Aug 2015.
Maxa JL, Melton LB, Ogu CC, Sills MN, Limanni A. Rhabdomyolysis after concomitant use of cyclosporine, simvastatin, gemfibrozil, and itraconazole. Ann Pharmacother. 2002;36(5):820–3.
Gottlieb A, Hoehndorf R, Dumontier M, Altman RB. Ranking adverse drug reactions with crowdsourcing. J Med Internet Res. 2015;17(3):e80.
Scheife R, Hines L, Boyce R, Chung S, Momper J, Sommer C, et al. Consensus recommendations for systematic evaluation of drug–drug interaction evidence for clinical decision support. Drug Saf. 2015;38(2):197–206.
Boyce RD, Ryan PB, Norén GN, Schuemie MJ, Reich C, Duke J, et al. Bridging islands of information to establish an integrated knowledge base of drugs and health outcomes of interest. Drug Saf. 2014;37(8):557–67.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported in part by National Library of Medicine (NLM) grant R01 LM011369, National Institute of General Medical Sciences (NIGMS) grant R01 GM101430, and National Human Genome Research Institute (NHGRI) grant U54 HG004028 for the National Center for Biomedical Ontology (NCBO).
Conflict of interest
Howard R. Strasberg is an employee of Wolters Kluwer Health; George Hripcsak has served as PI on grants to Columbia University from Janssen Research and from AstraZeneca on observational medical research, but there is no conflict of interest associated with the paper’s results. Juan M. Banda, Alison Callahan, Rainer Winnenburg, Aurel Cami, Ben Y. Reis, Santiago Vilar, Michel Dumontier, and Nigam Haresh Shah have no conflicts of interest that are directly relevant to the content of this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Banda, J.M., Callahan, A., Winnenburg, R. et al. Feasibility of Prioritizing Drug–Drug-Event Associations Found in Electronic Health Records. Drug Saf 39, 45–57 (2016). https://doi.org/10.1007/s40264-015-0352-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-015-0352-2