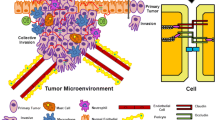
Abstract
The epithelial linings of eukaryotic organs form dynamically-regulated selectively-permeable barriers that control the movement of substances into (and out of) mucosal tissues. The principal structural determinants of epithelial barrier function are intercellular tight junctions (TJs), multi-protein complexes composed of claudin and non-claudin transmembrane proteins in addition to cytosolic linker proteins. As well as their crucial roles in barrier function, it is now well recognized that TJ proteins coordinate a variety of signaling and trafficking functions regulating physiological events such as cell differentiation, proliferation, migration and polarity. Accordingly, dysregulations in TJ protein expression or function are increasingly being linked to several pathophysiologies including cancer. To date, claudins have received the most attention as putative contributors to cancer initiation or progression. However the contribution of non-claudin transmembrane TJ proteins (including select immunoglobulin superfamily members, nectins, occludin and Marvel D family members) to the pathophysiology of cancer remains incompletely understood. Therefore the focus of this review is to collate recently-published evidence that supports or discounts a role for non-claudin transmembrane TJ proteins in cancer, and to speculate upon the feasibility of these molecules as prognostic biomarkers or therapeutic targets in cancer.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Gumbiner BM (1996) Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84(3):345–357
Leech AO, Cruz RG, Hill AD, Hopkins AM (2015) Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann Transl Med 3(13):184
Garrido-Urbani S, Bradfield PF, Imhof BA (2014) Tight junction dynamics: the role of junctional adhesion molecules (JAMs). Cell Tissue Res 355(3):701–715
Mandell KJ, Parkos CA (2005) The JAM family of proteins. Adv Drug Deliv Rev 57(6):857–867
Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y (2003) JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol 23(12):4267–4282
Arrate MP, Rodriguez JM, Tran TM, Brock TA, Cunningham SA (2001) Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J Biol Chem 276(49):45826–45832
Palmeri D, van Zante A, Huang CC, Hemmerich S, Rosen SD (2000) Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J Biol Chem 275(25):19139–19145
Aurrand-Lions M, Johnson-Leger C, Wong C, Du Pasquier L, Imhof BA (2001) Heterogeneity of endothelial junctions is reflected by differential expression and specific subcellular localization of the three JAM family members. Blood 98(13):3699–3707
Kobayashi I, Kobayashi-Sun J, Kim AD, Pouget C, Fujita N, Suda T et al (2014) Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512(7514):319–323
Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA et al (2007) JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med 204(13):3067–3076
Naik MU, Stalker TJ, Brass LF, Naik UP (2012) JAM-A protects from thrombosis by suppressing integrin alphaIIbbeta3-dependent outside-in signaling in platelets. Blood 119(14):3352–3360
Naik MU, Caplan JL, Naik UP (2014) Junctional adhesion molecule-A suppresses platelet integrin alphaIIbbeta3 signaling by recruiting Csk to the integrin-c-Src complex. Blood 123(9):1393–1402
Karshovska E, Zhao Z, Blanchet X, Schmitt MM, Bidzhekov K, Soehnlein O et al (2015) Hyperreactivity of junctional adhesion molecule A-deficient platelets accelerates atherosclerosis in hyperlipidemic mice. Circ Res 116(4):587–599
McSherry EA, McGee SF, Jirstrom K, Doyle EM, Brennan DJ, Landberg G et al (2009) JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer 125(6):1343–1351
•• Brennan K, McSherry EA, Hudson L, Kay EW, Hill AD, Young LS et al. (2013) Junctional adhesion molecule-A is co-expressed with HER2 in breast tumors and acts as a novel regulator of HER2 protein degradation and signaling. Oncogene 32(22):2799–804. In breast cancer tissue specimens, this manuscript demonstrated high JAM-A expression to correlate significantly with HER2 protein expression, ER negativity, lower patient age, high-grade breast cancers, and aggressive luminal B, HER2-positive and basal subtypes of breast cancer. Under in vitro conditions it demonstrated a novel putative regulation of HER2 expression by JAM-A in breast cancer cells
Murakami M, Giampietro C, Giannotta M, Corada M, Torselli I, Orsenigo F et al (2011) Abrogation of junctional adhesion molecule-A expression induces cell apoptosis and reduces breast cancer progression. PLoS One 6(6):e21242
•• Goetsch L, Haeuw JF, Beau-Larvor C, Gonzalez A, Zanna L, Malissard M et al. (2013) A novel role for junctional adhesion molecule-A in tumor proliferation: modulation by an anti-JAM-A monoclonal antibody. Int J Cancer 132(6):1463–1474. An antagonistic monoclonal antibody recognizing human JAM-A was developed and found to have significant anti-cancer properties in vivo in pre-clinical murine models of xenografted human breast tumors
•• Murakami M, Francavilla C, Torselli I, Corada M, Maddaluno L, Sica A et al. (2010) Inactivation of junctional adhesion molecule-A enhances antitumoral immune response by promoting dendritic cell and T lymphocyte infiltration. Cancer Res 70(5):1759–1765. This manuscript showed that growth and aggressiveness of a pancreatic islet cell carcinoma induced by SV40 T antigen expression in beta cells (Rip1Tag2 mice) was significantly attenuated in JAM-A-null mice, in conjunction with reduced angiogenesis and a marked increase in the immune response
Tian Y, Tian Y, Zhang W, Wei F, Yang J, Luo X et al (2015) Junctional adhesion molecule-A, an epithelial-mesenchymal transition inducer, correlates with metastasis and poor prognosis in human nasopharyngeal cancer. Carcinogenesis 36(1):41–48
Zhang M, Luo W, Huang B, Liu Z, Sun L, Zhang Q et al (2013) Overexpression of JAM-A in non-small cell lung cancer correlates with tumor progression. PLoS One 8(11):e79173
Ikeo K, Oshima T, Shan J, Matsui H, Tomita T, Fukui H et al (2015) Junctional adhesion molecule-A promotes proliferation and inhibits apoptosis of gastric cancer. Hepatogastroenterology 62(138):540–545
•• Lathia JD, Li M, Sinyuk M, Alvarado AG, Flavahan WA, Stoltz K et al. (2014) High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep 6(1):117–129. Demonstrated the importance of JAM-A for self renewal of cancer stem cells and growth of brain tumors
•• Thiagarajan PS, Hitomi M, Hale JS, Alvarado AG, Otvos B, Sinyuk M et al. (20105) Development of a fluorescent reporter system to delineate cancer stem cells in triple-negative breast cancer. Stem Cells 33(7):2114–2125. First evidence that JAM-A is highly expressed in, and is essential for, self renewal of breast cancer stem cells
•• Kelly KR, Espitia CM, Zhao W, Wendlandt E, Tricot G, Zhan F, et al. (2015) Junctional adhesion molecule-A is overexpressed in advanced multiple myeloma and determines response to oncolytic reovirus. Oncotarget 6(38):41275–41289. Demonstrated JAM-A as a predictive biomarker for sensitivity to Reolysin-induced cell death in multiple myeloma cells
Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP (2008) Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res 68(7):2194–2203
Wang Y, Lui WY (2012) Transforming growth factor-beta1 attenuates junctional adhesion molecule-A and contributes to breast cancer cell invasion. Eur J Cancer 48(18):3475–3487
Fong D, Spizzo G, Mitterer M, Seeber A, Steurer M, Gastl G et al (2012) Low expression of junctional adhesion molecule A is associated with metastasis and poor survival in pancreatic cancer. Ann Surg Oncol 19(13):4330–4336
Gutwein P, Schramme A, Voss B, Abdel-Bakky MS, Doberstein K, Ludwig A et al (2009) Downregulation of junctional adhesion molecule-A is involved in the progression of clear cell renal cell carcinoma. Biochem Biophys Res Commun 380(2):387–391
Coyne CB, Bergelson JM (2005) CAR: a virus receptor within the tight junction. Adv Drug Deliv Rev 57(6):869–882
Sollerbrant K, Raschperger E, Mirza M, Engstrom U, Philipson L, Ljungdahl PO et al (2003) The Coxsackievirus and adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein ligand-of-numb protein-X (LNX). J Biol Chem 278(9):7439–7444
Zhang Y, Bergelson JM (2005) Adenovirus receptors. J Virol 79(19):12125–12131
Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS et al (1997) Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275(5304):1320–1323
•• Reeh M, Bockhorn M, Gorgens D, Vieth M, Hoffmann T, Simon R et al. (2013) Presence of the coxsackievirus and adenovirus receptor (CAR) in human neoplasms: a multitumour array analysis. Br J Cancer 109(7):1848–1858. Examined differential expression of CAR in human neoplasms across many types of cancers and made interesting conclusions surrounding potential hormonal effects due to contrasting CAR expression in female versus male reproductive organs. Additionally, this study established novel CAR expression levels in specific cancers including basalioma, warthin’s tumour and thyroid adenoma
Giaginis CT, Zarros AC, Papaefthymiou MA, Papadopouli AE, Sfiniadakis IK, Theocharis SE (2008) Coxsackievirus and adenovirus receptor expression in human endometrial adenocarcinoma: possible clinical implications. World J Surg Oncol 6:59
Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT (2000) The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res 60(18):5031–5036
Qin M, Escuadro B, Dohadwala M, Sharma S, Batra RK (2004) A novel role for the coxsackie adenovirus receptor in mediating tumor formation by lung cancer cells. Cancer Res 64(18):6377–6380
Veena MS, Qin M, Andersson A, Sharma S, Batra RK (2009) CAR mediates efficient tumor engraftment of mesenchymal type lung cancer cells. Lab Investig 89(8):875–886
• Chen Z, Wang Q, Sun J, Gu A, Jin M, Shen Z et al. (2013) Expression of the coxsackie and adenovirus receptor in human lung cancers. Tumour Biol 34(1):17–24. Demonstrated upregulation of CAR in lung cancer tumour vs normal tissues. Demonstrated how silencing CAR could inhibit colony formation, invasive phenotype and adhesive nature of A549 cells compared to CAR overexpression. An important demonstration of how targeting CAR may reduce tumorigenic phenotypes of cancers featuring its overexpression
Vindrieux D, Le Corre L, Hsieh JT, Metivier R, Escobar P, Caicedo A et al (2011) Coxsackie and adenovirus receptor is a target and a mediator of estrogen action in breast cancer. Endocr Relat Cancer 18(3):311–321
Martin TA, Watkins G, Jiang WG (2005) The Coxsackie-adenovirus receptor has elevated expression in human breast cancer. Clin Exp Med 5(3):122–128
Wunder T, Schumacher U, Friedrich RE (2012) Coxsackie adenovirus receptor expression in carcinomas of the head and neck. Anticancer Res 32(3):1057–1062
Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V et al (2002) Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res 62(13):3812–3818
Anders M, Vieth M, Rocken C, Ebert M, Pross M, Gretschel S et al (2009) Loss of the coxsackie and adenovirus receptor contributes to gastric cancer progression. Br J Cancer 100(2):352–359
Sachs MD, Rauen KA, Ramamurthy M, Dodson JL, De Marzo AM, Putzi MJ et al (2002) Integrin alpha(v) and coxsackie adenovirus receptor expression in clinical bladder cancer. Urology 60(3):531–536
Buscarini M, Quek ML, Gilliam-Hegarich S, Kasahara N, Bochner B (2007) Adenoviral receptor expression of normal bladder and transitional cell carcinoma of the bladder. Urol Int 78(2):160–166
Kuster K, Koschel A, Rohwer N, Fischer A, Wiedenmann B, Anders M (2010) Downregulation of the coxsackie and adenovirus receptor in cancer cells by hypoxia depends on HIF-1alpha. Cancer Gene Ther 17(2):141–146
Stecker K, Vieth M, Koschel A, Wiedenmann B, Rocken C, Anders M (2011) Impact of the coxsackievirus and adenovirus receptor on the adenoma-carcinoma sequence of colon cancer. Br J Cancer 104(9):1426–1433
Wold WS, Toth K (2013) Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther 13(6):421–433
Takai Y, Miyoshi J, Ikeda W, Ogita H (2008) Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol 9(8):603–615
Izumi H, Hirabayashi K, Nakamura N, Nakagohri T (2015) Nectin expression in pancreatic adenocarcinoma: nectin-3 is associated with a poor prognosis. Surg Today 45(4):487–494
Takai Y, Nakanishi H (2003) Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci 116(Pt 1):17–27
Samanta D, Almo SC (2015) Nectin family of cell-adhesion molecules: structural and molecular aspects of function and specificity. Cell Mol Life Sci 72(4):645–658
Elloul S, Kedrin D, Knoblauch NW, Beck AH, Toker A (2014) The adherens junction protein afadin is an AKT substrate that regulates breast cancer cell migration. Mol Cancer Res 12(3):464–476
Fournier G, Cabaud O, Josselin E, Chaix A, Adelaide J, Isnardon D et al (2011) Loss of AF6/afadin, a marker of poor outcome in breast cancer, induces cell migration, invasiveness and tumor growth. Oncogene 30(36):3862–3874
Sun TT, Wang Y, Cheng H, Xiao HZ, Xiang JJ, Zhang JT et al (2014) Disrupted interaction between CFTR and AF-6/afadin aggravates malignant phenotypes of colon cancer. Biochim Biophys Acta 1843(3):618–628
Yamamoto T, Mori T, Sawada M, Matsushima H, Ito F, Akiyama M et al (2015) Loss of AF-6/afadin induces cell invasion, suppresses the formation of glandular structures and might be a predictive marker of resistance to chemotherapy in endometrial cancer. BMC Cancer 15:275
• Xu Y, Chang R, Peng Z, Wang Y, Ji W, Guo J et al (2015) Loss of polarity protein AF6 promotes pancreatic cancer metastasis by inducing Snail expression. Nat Commun 6:7184. Showed evidence of AF6 as a potential inhibitor of metastasis in pancreatic cancer cells
Martin TA, Lane J, Harrison GM, Jiang WG (2013) The expression of the Nectin complex in human breast cancer and the role of Nectin-3 in the control of tight junctions during metastasis. PLoS One 8(12):e82696
Oshima T, Sato S, Kato J, Ito Y, Watanabe T, Tsuji I et al (2013) Nectin-2 is a potential target for antibody therapy of breast and ovarian cancers. Mol Cancer 12:60
Karabulut M, Gunaldi M, Alis H, Afsar CU, Karabulut S, Serilmez M et al (2015) Serum nectin-2 levels are diagnostic and prognostic in patients with colorectal carcinoma. Clin Transl Oncol 18(2):160–171
Miao X, Yang ZL, Xiong L, Zou Q, Yuan Y, Li J et al. (2013) Nectin-2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol 6(2):179–190
Nabih ES, Abdel Motaleb FI, Salama FA (2014) The diagnostic efficacy of nectin 4 expression in ovarian cancer patients. Biomarkers 19(6):498–504
Derycke MS, Pambuccian SE, Gilks CB, Kalloger SE, Ghidouche A, Lopez M et al (2010) Nectin 4 overexpression in ovarian cancer tissues and serum: potential role as a serum biomarker. Am J Clin Pathol 134(5):835–845
Fabre-Lafay S, Monville F, Garrido-Urbani S, Berruyer-Pouyet C, Ginestier C, Reymond N et al (2007) Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer 7:73
Takano A, Ishikawa N, Nishino R, Masuda K, Yasui W, Inai K et al (2009) Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res 69(16):6694–6703
Das D, Satapathy SR, Siddharth S, Nayak A, Kundu CN (2015) NECTIN-4 increased the 5-FU resistance in colon cancer cells by inducing the PI3K-AKT cascade. Cancer Chemother Pharmacol 76(3):471–479
Lattanzio R, Ghasemi R, Brancati F, Sorda RL, Tinari N, Perracchio L et al (2014) Membranous Nectin-4 expression is a risk factor for distant relapse of T1-T2, N0 luminal-A early breast cancer. Oncogenesis 3:e118
Raveh S, Gavert N, Spiegel I, Ben-Ze’ev A (2009) The cell adhesion nectin-like molecules (Necl) 1 and 4 suppress the growth and tumorigenic ability of colon cancer cells. J Cell Biochem 108(1):326–336
Rao R (2009) Occludin phosphorylation in regulation of epithelial tight junctions. Ann N Y Acad Sci 1165:62–68
Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S et al (1993) Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 123(6 Pt 2):1777–1788
Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S et al (1994) Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol 127(6 Pt 1):1617–1626
Dorfel MJ, Huber O (2012) Modulation of tight junction structure and function by kinases and phosphatases targeting occludin. J Biomed Biotechnol 2012:807356
Cummins PM (2012) Occludin: one protein, many forms. Mol Cell Biol 32(2):242–250
Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H et al (2010) The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev Cell 18(1):52–63
Martin TA, Mansel RE, Jiang WG (2010) Loss of occludin leads to the progression of human breast cancer. Int J Mol Med 26(5):723–734
Hwang I, An BS, Yang H, Kang HS, Jung EM, Jeung EB (2013) Tissue-specific expression of occludin, zona occludens-1, and junction adhesion molecule A in the duodenum, ileum, colon, kidney, liver, lung, brain, and skeletal muscle of C57BL mice. J Physiol Pharmacol 64(1):11–18
Tokunaga Y, Tobioka H, Isomura H, Kokai Y, Sawada N (2004) Expression of occludin in human rectal carcinoid tumours as a possible marker for glandular differentiation. Histopathology 44(3):247–250
Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N (2006) Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res 66(18):9125–9133
Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N (2007) Occludin-mediated premature senescence is a fail-safe mechanism against tumorigenesis in breast carcinoma cells. Cancer Sci 98(7):1027–1034
Beeman N, Webb PG, Baumgartner HK (2012) Occludin is required for apoptosis when claudin-claudin interactions are disrupted. Cell Death Dis 3:e273
• Rachow S, Zorn-Kruppa M, Ohnemus U, Kirschner N, Vidal-y-Sy S, von den Driesch P et al (2013) Occludin is involved in adhesion, apoptosis, differentiation and Ca2 + -homeostasis of human keratinocytes: implications for tumorigenesis. PLoS One 8(2):e55116. Demonstrated a role for occludin in apoptotic signaling, when its silencing reduced susceptibility to TRAIL in human keratinocytes. Important illustration of a potential mechanism by which altered occludin expression could drive tumorigenesis
Runkle EA, Rice SJ, Qi J, Masser D, Antonetti DA, Winslow MM et al (2012) Occludin is a direct target of thyroid transcription factor-1 (TTF-1/NKX2-1). J Biol Chem 287(34):28790–28801
Cichon C, Sabharwal H, Ruter C, Schmidt MA (2014) MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2(4):e944446
Ye D, Guo S, Al-Sadi R, Ma TY (2011) MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 141(4):1323–1333
Li D, Mrsny RJ (2000) Oncogenic Raf-1 disrupts epithelial tight junctions via downregulation of occludin. J Cell Biol 148(4):791–800
Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S (2005) Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171(6):939–945
Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K et al (2006) Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet 79(6):1040–1051
Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S (2008) Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell 19(11):4687–4693
Sanchez-Pulido L, Martin-Belmonte F, Valencia A, Alonso MA (2002) MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci 27(12):599–601
Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y et al (2010) Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 21(7):1200–1213
• Oda Y, Otani T, Ikenouchi J, Furuse M (2014) Tricellulin regulates junctional tension of epithelial cells at tricellular contacts through Cdc42. J Cell Sci 127(Pt 19):4201–4212. This manuscript highlighted for the first time the contribution of tricellular contacts to junctional tension and cellular shape in epithelial cells
Kojima T, Fuchimoto J, Yamaguchi H, Ito T, Takasawa A, Ninomiya T et al (2010) c-Jun N-terminal kinase is largely involved in the regulation of tricellular tight junctions via tricellulin in human pancreatic duct epithelial cells. J Cell Physiol 225(3):720–733
Kojima T, Takasawa A, Kyuno D, Ito T, Yamaguchi H, Hirata K et al (2011) Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells. Exp Cell Res 317(16):2288–2298
Korompay A, Borka K, Lotz G, Somoracz A, Torzsok P, Erdelyi-Belle B et al (2012) Tricellulin expression in normal and neoplastic human pancreas. Histopathology 60(6B):E76–E86
Somoracz A, Korompay A, Torzsok P, Patonai A, Erdelyi-Belle B, Lotz G et al (2014) Tricellulin expression and its prognostic significance in primary liver carcinomas. Pathol Oncol Res 20(4):755–764
Patonai A, Erdelyi-Belle B, Korompay A, Somoracz A, Straub BK, Schirmacher P et al (2011) Claudins and tricellulin in fibrolamellar hepatocellular carcinoma. Virchows Arch 458(6):679–688
Masuda R, Semba S, Mizuuchi E, Yanagihara K, Yokozaki H (2010) Negative regulation of the tight junction protein tricellulin by snail-induced epithelial-mesenchymal transition in gastric carcinoma cells. Pathobiology 77(2):106–113
Kondoh A, Takano K, Kojima T, Ohkuni T, Kamekura R, Ogasawara N et al (2011) Altered expression of claudin-1, claudin-7, and tricellulin regardless of human papilloma virus infection in human tonsillar squamous cell carcinoma. Acta Otolaryngol 131(8):861–868
Steed E, Rodrigues NT, Balda MS, Matter K (2009) Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC cell biology. 10:95
• Cording J, Berg J, Kading N, Bellmann C, Tscheik C, Westphal JK et al. (2013) In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci 126(Pt 2):554–564. This paper provided in-depth evidence of the interaction of tight-junction-associated marvel proteins (TAMPs) occludin, tricellulin and MarvelD3 with claudins
Tessema M, Yingling CM, Liu Y, Tellez CS, Van Neste L, Baylin SS et al (2014) Genome-wide unmasking of epigenetically silenced genes in lung adenocarcinoma from smokers and never smokers. Carcinogenesis 35(6):1248–1257
• Steed E, Elbediwy A, Vacca B, Dupasquier S, Hemkemeyer SA, Suddason T et al. (2014) MarvelD3 couples tight junctions to the MEKK1-JNK pathway to regulate cell behavior and survival. J Cell Biol 204(5):821–838. This paper identified a novel role for MarvelD3 in linking tight junctions with the MEKK1-JNK pathway, thereby regulating cellular functions of importance in tumorigenesis
Acknowledgments
We thank current and former members of the Hopkins laboratory for their hard work and helpful discussions, and are grateful to the following funding agencies for support in recent years: Science Foundation Ireland (13/IA/1994; 2008/RFP/NSC1427; 2008/RFP/NSC1427 TIDA Feasibility 10, to AMH); Health Research Board of Ireland (HRA POR-2014-545; HRA/2009/49; RP/2006/95, to AMH); Breast Cancer Ireland; The Beaumont Hospital Cancer Research and Development Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Sri HariKrishna Vellanki, Cathy E Richards, Yvonne E Smith, and Ann M Hopkins declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
Among cited articles where one of the authors of the current report were authors, local Institutional Review Board approval was obtained and maintained for studies where human (or animal) subjects research was performed.
Additional information
This article is part of the Topical Collection on Leaky Junctions in Cancer.
Rights and permissions
About this article
Cite this article
Vellanki, S.H., Richards, C.E., Smith, Y.E. et al. The Contribution of Ig-Superfamily and MARVEL D Tight Junction Proteins to Cancer Pathobiology. Curr Pathobiol Rep 4, 37–46 (2016). https://doi.org/10.1007/s40139-016-0105-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40139-016-0105-7