Abstract
Purpose
Rare yeasts species are increasingly reported as causative agents of invasive human infection. Proper identification and antifungal therapy are essential to manage these infections. Candida blankii is one of these emerging pathogens and is known for its reduced susceptibility to multiple antifungals.
Methods
To obtain more insight into the characteristics of this species, 26 isolates reported as C. blankii were investigated using genetic and phenotypical approaches.
Results
Among the 26 isolates, seven recovered either from blood, sputum, urine, or the oral cavity, displayed substantial genetic and some phenotypical differences compared to the other isolates, which were confirmed as C. blankii. We consider these seven strains to represent a novel species, Tardiomyces depauwii. Phylogenomics assigned C. blankii, C. digboiensis, and the novel species in a distinct branch within the order Dipodascales, for which the novel genus Tardiomyces is erected. The new combinations Tardiomyces blankii and Tardiomyces digboiensis are introduced. Differences with related, strictly environmental genera Sugiyamaella, Crinitomyces, and Diddensiella are enumerated. All three Tardiomyces species share the rare ability to grow up to 42 °C, display slower growth in nutrient-poor media, and show a reduced susceptibility to azoles and echinocandins. Characteristics of T. depauwii include high MIC values with voriconazole and a unique protein pattern.
Conclusion
We propose the novel yeast species Tardiomyces depauwii and the transfer of C. blankii and C. digboiensis to the novel Tardiomyces genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yeast species are among the primary causative agents of invasive fungal infections. In recent years, invasive infections due to less common species are increasingly reported, partially due to advances in molecular diagnostics and an increase in vulnerable hosts [1,2,3]. Some of these emerging species pose a threat to public health due to difficulties in identification, limited therapeutic options caused by resistance and the risk of nosocomial outbreaks with undetermined sources of infection [4,5,6,7]. Originally described in the organs of a mink in 1968 [8], Candida blankii is considered an emerging human pathogenic fungus that has been isolated from varying environments like the surface of apples, flowers, and was the most abundant species in surface coastal marine habitats in Vietnam [9,10,11]. Environmental C. blankii isolates have been used in biotechnological research [12]. The first clinical isolate of C. blankii was reported in a cystic fibrosis (CF) patient in 2015 [13]. Since then, C. blankii infections were reported worldwide in patients with endocarditis, cystic fibrosis (CF), SARS-CoV-2, and in a preterm neonate [14,15,16,17,18]. Furthermore, C. blankii was the causative agent of an outbreak in a neonatal intensive care unit in Delhi, India, which resulted in bloodstream infections in nine neonates over a seven-month period [19]. Additional clinical isolates were retrospectively identified in surveillance studies in Norway and the United Kingdom, albeit in low numbers [20, 21]. A shared characteristic among all investigated strains is the reduced susceptibility against various azoles and echinocandins, making this species difficult to manage. Identification via phenotypic methods appeared challenging, and therefore molecular identification methods are preferred [16].
The closest relative of C. blankii is C. digboiensis, isolated from sludge-contaminated soil, tar and acid mine drainage in Zambia and India [22,23,24]. Recently, C. digboiensis was reported as the causative agent of candidemia in the United Kingdom [25]. ITS rDNA sequences of both species clustered them in a distinct branch, distant from the CTG clade that includes common Candida species like Candida albicans, Candida tropicalis, and Candida parapsilosis [14].
In the present study, we demonstrate that several European clinical isolates, previously reported as C. blankii, were found to harbor genomic and phenotypical differences to isolates curated under the same name, while demonstrating minimal genetic diversity between each other. For the latter cohesive group, we propose a novel yeast species, closely related to C. blankii. Given the large phylogenetic distance of both species to C. albicans, a model type of the genus Candida, we propose the novel genus Tardiomyces to accommodate this yeast. With this naming, the reassignment of the previously known C. blankii and C. digboiensis to the genus Tardiomyces, thus, is necessary.
Materials and methods
Isolate culture and DNA extraction
A total of 26 isolates initially reported as C. blankii were cultured for two days on Sabouraud dextrose agar (SDA) (Oxoid, Hampshire, United Kingdom) at 30 °C after being taken from storage at −80 °C (Table S1). C. albicans American Type Culture Collection (ATCC) 10231, C. tropicalis ATCC 750T, C. parapsilosis ATCC 22019T, Pichia kudriavzevii (Candida krusei) ATCC 6258T, and Nakaseomyces glabratus (Candida glabrata) ATCC 90030 were used in each experiment as controls. For each isolate, a colony was randomly selected and resuspended in 700 µL MagNA Pure Bacteria lysis buffer and MagNA Lyser green beads and mechanically lysed for 30 s at 6500 rpm with the MagNA Lyser system (All Roche Diagnostics GmbH, Mannheim, Germany). DNA was extracted and purified with the MagNA Pure 96 instrument with the MagNA Pure DNA and Viral Small Volume Kit (Roche Diagnostics), following the manufacturer’s instruction.
Identification methods
Isolates were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) using a Bruker MALDI Biotyper system as described [26]. Biochemical profile analysis with a VITEK 2XL (BioMérieux, Marcy l’Etoile, France) was done according to manufacturer’s instruction using VITEK 2 YST ID card and VITEK 2XL identification software v9.02 and were analyzed in triplicates. For internal transcribed spacers (ITS) sequencing, ITS-1 and ITS-4 primers were used as described [27]. Control sequences were extracted from the National Center for Biotechnology Information (NCBI) nucleotide database and alignment was generated using MAFFT v7 [28], and the phylogenetic tree was built with IQ-TREE web server with default parameters [29] (Table S2). Visualization and edition were made using iTOL v6 [30]. ITS sequences generated in the current study were deposited to the NCBI Genbank database (Accession numbers: OR479767–OR479792).
Whole-genome sequencing (WGS)
For WGS, DNA was extracted as described above and samples were subsequently treated with RNase A (Merck KGaA, Darmstadt, Germany) at a final concentration of 5 µg/µL for one hour at room temperature. DNA was afterward extracted and purified again as before and then measured with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) using the double-stranded DNA (dsDNA) high-sensitivity option. For Illumina sequencing, genomic libraries were prepared and sequenced with the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA) with 2-by-150 bp paired-end-read mode at Eurofins Genomics (Ebersberg, Germany). For Oxford Nanopore sequencing, libraries were prepared using the ONT rapid sequencing kit and were sequenced using a GridION (Oxford Nanopore Technologies [ONT], Oxford, UK) instrument. Raw read data generated in this study have been submitted to the NCBI Sequence Read Archive (BioProject ID: PRJNA1009266).
Assembly and annotation
Generated Nanopore and Illumina reads were uploaded to the Galaxy platform and were assessed with FastQC [31]. De novo assembly was performed on Nanopore reads with Flye v2.9.1 using the Nanopore raw mode [32]. Illumina reads of the same strain were aligned to the generated assembly using BWA-MEM v0.7.17, PCR duplicates were removed with RmDup, local realignment was performed using BamLeftAlign and unpaired reads were removed with BAM filter [33]. The draft assembly was improved using the generated BAM file and Pilon v.1.20.1 [34]. Gene prediction and annotation were performed using Funannotate pipeline v1.8.15 [35], including soft masking with RepeatMasker and RepeatModeler tools, ab initio training using Augustus, Snap, glimmerHMM, codingQuary, and GenMark. Functional annotation was assigned by similarity using Pfam, MEROPS, CAZy, eggNOG, InterProScan, Antismash, SignalP, and Swissprot databases by BLASTP or HMMER3 searches using Funannotate default parameters. Genome annotations are available online (Supplementary table). Candidate heterozygous positions predicted by Pilon v1.20.1 were examined using JBrowse v1.16.11 [36]. For de novo assembly based on Illumina reads, SPAdes v3.15.4 with default settings was used except for the assembly and error correction operation mode and paired-end: individual datasets reads options [37]. Genome assembly qualities was assessed with Quast v5.2.0 [37]. The generated assembly was deposited to the NCBI Assembly database (BioProject ID: PRJNA1009997).
Phylogenomic analysis and comparative genomics
Orthology of 39 species of Saccharomycotina subphylum was accessed using OrthoFinder v2.5.4 [38], with default parameters to identify orthologous gene groups and Multiple Sequence Alignment option to infer a rooted species tree (Table S3). Schizosaccharomyces pombe and Cryptococcus neoformans were used as outgroup species. The generated tree was visualized and edited using iTol v6 [30]. Orthologous gene clusters of Tardiomyces depauwii, T. blankii, T. digboiensis, and two closely related species Crinitomyces ghanaensis and Sugiyamaella lignohabitans were compared using OrthoVenn2 using an e-value cutoff of 1 × 10–5 and inflation values of 1.5 [39].
SNPs in T. depauwii isolates were called using Freebayes v.1.3.6 as previously described using a validated pipeline [40]. In short, SNPs in the resulting VCF file with a read depth (DP) of < 15, a quality of < 100 and an allele frequency between 0.15 and 0.90 × DP were removed. Phylogenetic analysis and visualization were performed using VCF2PopTree, MEGA11 v11.0.10 and iTOL v6 as previously described [41].
Bayesian phylogenies were generated with BEAST v.1.10.4 [42]. The filtered VCF file was converted to FASTA alignments with vcf2phylip v2.8 and was subsequently converted to an xml file with BEAUti v1.10.4 [43]. Random nucleotides totaling the T. depauwii genomic size minus the number of positions on the VCF file were inserted in the generated xml file. Tip dates were set to the year of collection. The GTR model with gamma variation among sites was used as the substitution model with a strict molecular clock. A coalescent exponential growth model was selected as the Tree Prior and the length of chain was set to 50,000,000. The generated log file was inspected with Tracer v1.7.2, annotation was performed with TreeAnnotator v1.10.4 with the burnin (as states) set to 5,000,000 and subsequently visualized with FigTree v1.4.4 [44].
Phenotypic characterization
Micro- and macroscopic morphology were determined after 48 h of incubation on yeast peptone dextrose (YPD) plates (Thermo Fisher Scientific) and CandiSelect Agar (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 30 °C. Physiological characterization of temperature effects, osmotolerance, NaCl stress, and urease hydrolysis on growth was performed using standard procedures described in [45]. API 20 C AUX (BioMérieux), strips were used as suggested by the manufacturer and incubated at 30 °C for 72 h. Assimilation patterns were read after 72 h of incubation. For measuring growth rates of T. depauwii, T. blankii, T. digboiensis, and common yeast species (C. albicans, C. tropicalis, C. parapsilosis, P. kudriavzevii, N. glabratus), cell suspensions were diluted in water to a final concentration of 1–5 × 103 CFU/mL using a Genesys 20 Spectrometer (Thermo Fisher Scientific). Cultures were incubated at 35 °C and growth was quantitated by measuring optical density (OD) at 450 nm, using an 800 TS Microplate Reader (BioTek, Winooski, VA, USA). Isolates were incubated in YPD broth (Thermo Fisher Scientific), SAB (Oxoid) and RPMI 1640 medium with 0.2% and also 2% glucose (Thermo Fisher Scientific). The experiment was replicated eight times and averages with standard error of the mean were calculated using Prism v8.4.3 (Boston, MA, USA).
Antifungal susceptibility testing (AFST)
In vitro AFST in accordance with the Clinical & Laboratory Standards Institute (CLSI) M27 standard was performed against eight common antifungals [46]. In short, isolates were diluted to obtain a final concentration of 1–5 × 103 CFU-mL in RPMI 1640 medium (Thermo Fisher Scientific). Microtiter plates were incubated at 35 °C and visually read after 24 and 48 h as the first reading showed inadequate growth. Minimal inhibitory concentration (MIC) values were determined as the lowest antifungal concentration with a 50% growth reduction when compared to the growth control, except amphotericin B with a 100% growth reduction. Tested concentrations for all drugs ranged between 0.016 and 16 µg/mL, except for fluconazole with a range of 0.063–64 µg/mL. The Sensititre YeastOne (SYO) YO10 (Thermo Fisher Scientific) AFST was performed following manufacturer’s recommendations. Microtiter plates were incubated at 35 °C for 48 h, as incubation after 24 h did not yield sufficient growth, and SYO colorimetric MICs were recorded as the first well showing a distinct color change as compared to the positive control. AFST by E-test for amphotericin B was performed for all T. depauwii isolates. Inoculum suspensions of 0.5 McFarland were inoculated on the entire surface of the RPMI plate with a sterile cotton swab. E-test strips (Liofilchem, Roseto degli Abruzzi, Italy) were placed on the center of the RPMI 1640 plate and incubated at 35 °C for 72 h. The MIC was determined from the inhibition ellipse that intersected with the scale of the strip after 48 and 72 h as 24 h of incubation showed not enough growth.
Results
Isolate identification
All isolates, reported previously as C. blankii, were investigated by means of MALDI-TOF MS, ITS sequencing and the VITEK 2XL system. We also included a single C. digboiensis strain (type strain CBS 9800), the most related species to C. blankii. Out of 26 isolates, 19 were identified as C. blankii by MALDI-TOF MS, with score values of ≥ 1.7, and a 100% ITS sequence similarity match to multiple annotated C. blankii strains submitted to the NCBI Nucleotide database, also confirmed by phylogenetic analysis (Fig. 1A and B). The remaining seven strains could not be identified with MALDI-TOF MS, and ITS analysis displayed a difference of 21 substitutions and two indels from other C. blankii. For CBS 9800, identification using MALDI-TOF MS was also not possible. VITEK 2XL results of all isolates yielded either Cryptococcus laurentii (n = 13), Candida ciferrii (n = 4), Clavispora (Candida) lusitaniae (n = 1), or unidentified species (n = 9).
Identification results according to ITS sequencing, MALDI-TOF MS and VITEK. A MALDI-TOF MS and VITEK analysis were used to identify the isolates. *, CBS 9800 is C. digboiensis and included as control. B Phylogenetic tree of species from the Dipodascales order based on ITS sequences. The tree is constructed with maximum likelihood implemented in IQ-TREE web server using TPM3u + F + G4 model. Ultrafast bootstrap was used to 1000 resampled data and values > 80 are shown. Strains retrieved from NCBI are indicated in bold. Candida albicans was taken as an outgroup
Phylogenetic placement
From the seven strains that were not identified as C. blankii with MALDI-TOF, strain NCPF13064 was used to generate a high quality de novo assembly using Nanopore reads with Illumina read correction. This assembly generated nine contigs, with lengths ranging from 12 Kbp to 3.82 Mbp, yielding a total length of 13.47 Mbp (N50 = 3,623,071 bp, GC% = 57.69%). Genome annotation yielded 4867 protein-coding genes. Inspection of potential heterozygous positions according to aligned Illumina reads did not reveal such positions, indicating a haploid genome. Using this strain and type strains of C. blankii, C. digboiensis, 36 other Saccharomycotina genomes and two outgroups, comparative genomics was used to construct a phylogenomic tree based on ortholog groups, which showed a congruent topology in Saccharomycotina, grouped within the order Dipodascales, indicating a close relation between strain NCPF13064 and C. blankii and a common ancestor together with C. digboiensis (Fig. 2A). Strain NCPF13064 was differentiated from C. blankii, at a similar distance as found between the sister species C. albicans and C. dubliniensis, demonstrating that this strain together with the other five related isolates constitute a different, previously unrecognized species. Bases on this, we propose to name this novel species as Tardiomyces depauwii. Isolates of this species were isolated in Europe (Norway, the United Kingdom and the Netherlands) and the United States (New York), between 1991 and 2021 (Fig. S1). Furthermore, we propose the reassignment of the related C. blankii and C. digboiensis to the genus Tardiomyces, as they are grouped with T. depauwii and differ from the other described Candida species. With comparative genomic analysis, T. depauwii exhibited nine unique genes in comparison to the other species. For T. blankii and T. digboiensis, this number was 22 and 126 respectively (Fig. 2B). The number of genes for species in the genus Tardiomyces ranged between 4377 and 4702 (Fig. 2C).
Phylogenetic analysis based on ortholog groups from Saccharomycotina including the Tardiomyces genus. A Species tree based on proteomes from Saccharomycotina built from 2843 orthologous groups of genes. Tardiomyces spp. are highlighted. Cryptococcus neoformans and Schizosaccharomyces pombe were taken as outgroups. B Venn diagram of T. depauwii, T. blankii, T. digboiensis, C. ghanaensis and S. lignohabitans showing the number of shared and unique gene clusters for each species. C Overview of the total number of genes for each species. *3205 shared genes. †83 shared genes
WGS SNP analysis was performed on all seven T. depauwii isolates. SNP differences ranged from 23 to 459 SNPs between strains on a genome of approximately 13.5 Mbp (Fig. 3A). Divergence times between isolates were estimated with Bayesian inference utilizing the GTR substitution model with a coalescent exponential growth. Using Tracer, ESS values were sufficient for all parameters. The estimated time to the most recent common ancestor (TMRCA) for these T. depauwii isolates was as recent as around 1971 (Fig. 3B). Finally, using this pipeline it was not possible to map most reads of the T. depauwii isolates on the C. blankii reference genome NCPF13064.
WGS SNP and molecular clock analysis on all T. depauwii strains. A WGS SNP analysis, the tree was generated with MEGA11 v11.10.11 using the neighbor-joining tree method. B Molecular clock analysis with estimated dates of the most recent common ancestor. The tree was generated with BEAST v.1.10.4 using a strict clock with a coalescent model
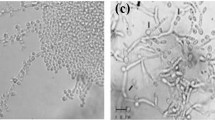
Phenotypic characterization
All seven T. depauwii strains formed smooth creamy white colored colonies after incubation for 48 h on YPD agar (Fig. 4A). Isolates propagated by means of budding cells, which were ellipsoidal to subspherical, 3–5 µm by 3–7 µm or via the formation of pseudohyphae (Fig. 4B). On CandiSelect Agar, colonies appeared light to dark blue (Fig. 4C). The six strains did not show any morphological differences between each other. Physiological characteristics regarding fermentation and assimilation of carbon sources, osmo- and saline tolerance, urease hydrolysis, and thermotolerance, were examined for all T. depauwii, T. blankii and T. digboiensis strains (Table 1). The fermentation and assimilation of carbon sources were highly similar between these species with only few differences. No phenotypic intra-species variation among T. depauwii strains was observed, while for the T. blankii strains five substrates were variably assimilated (Table S4). Phenotypical results for 16 substrates included in the API 20 C AUX confirmed the results mentioned above (Table S5). While T. blankii was able to grow in up to 60% glucose and 10% NaCl, T. depauwii and T. digboiensis did not show growth at 50% glucose and grew at a maximum saline concentration of 7.5%. All three investigated Tardiomyces species displayed growth up to 42 °C.
Growth profile
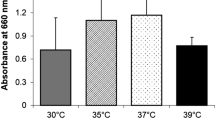
Growth rates of randomly selected T. depauwii (n = 3), T. blankii (n = 4), T. digboiensis (n = 1) and five other common human pathogenic yeast species (C. albicans, C. tropicalis, C. parapsilosis, P. kudriavzevii and N. glabratus) were investigated in RPMI 1640 medium (0.2% glucose), YPD broth and SAB broth. For RPMI 1640 with 0.2% glucose, all common yeast species reached the stationary phase around 24 h of incubation at 35 °C, while all isolates from the Tardiomyces genus exhibited a much slower growth rate, showing continuous growth till 48 h, with no indication of having reached the stationary phase (Fig. 5A). In YPD broth and SAB broth, all T. depauwii and T. digboiensis strains showed a greatly reduced growth rate when compared to the control strains, displaying continuous growth at 48 h, while most control strains reached the stationary phase around 24 h of incubation (Fig. 5B, C). Notably, the three T. blankii strains grew with the same rate as the common yeast species and one showed similar growth rates as T. depauwii and T. digboiensis (Fig. S2). To determine whether the higher glucose content of these media was involved, we also grew all strains in RPMI 1640 medium with 2% glucose (Fig. S3). Also, in this medium the same T. blankii strains demonstrated an elevated growth rate, as compared to the T. depauwii and T. digboiensis strains, however, they did not reach a growth rate as found for the common yeast strains (Fig. S2).
Antifungal susceptibility
In vitro AFST according to CLSI microbroth dilution was performed against eight common antifungals on all T. depauwii isolates (n = 7) and on T. blankii (n = 4) and T. digboiensis (n = 1) isolates as used before in the growth rate experiment. Due to minimal growth at 24 h of incubation, MICs values were read after 48 h of incubation. Notably, the T. depauwii strains demonstrated elevated fluconazole and voriconazole MICs of ≥ 64 µg/mL and 2–8 µg/mL, respectively (Table 2). For T. blankii, fluconazole MICs ranged from 8 to 32 µg/mL in the four tested strains, while the T. digboiensis isolate showed a MIC of 4 µg/mL. Anidulafungin and micafungin MICs were elevated for nearly all Tardiomyces strains, ranging mostly between 0.125 and 8 µg/mL. Overall, anidulafungin MICs were twofold dilutions higher than micafungin MICs for T. depauwii and T. blankii. Amphotericin B MICs were either 0.25 or 0.5 µg/mL for all tested Tardiomyces isolates. MIC values according to SYO were similar, confirming the elevated MICs for most azoles and echinocandins of all Tardiomyces strains and the higher MICs of anidulafungin when compared to micafungin. Finally, SYO demonstrated that flucytosine MICs were low, with MICs being ≤ 0.12 µg/mL (Table S6). Amphotericin B MICs for T. depauwii strains using E-tests were read after 48 and 72 h and ranged between 0.38 and 1 µg/mL, and 0.5 and 1 µg/mL, respectively (Table S7).
Discussion
In the present study, we described a novel yeast species and proposed the reassignment of C. blankii and C. digboiensis to a novel genus Tardiomyces. Genotypic analysis of the seven clinical T. depauwii isolates revealed an extremely low genetic diversity, despite originating from different countries during a 32-year period, and clear delineation from the related T. blankii (C. blankii) and T. digboiensis (C. digboiensis). The genus Tardiomyces is characterized by an overall low growth rate in comparison to common pathogenic yeast species, elevated MICs against azoles and echinocandins, especially against fluconazole, voriconazole and anidulafungin in most of the strains, and growth up to 42 °C.
Identification and genetics of Tardiomyces
Recent advances in molecular taxonomy have facilitated the increase of reported rare yeasts, and the description of novel species and genera [47]. Molecular identification methods like ITS sequencing or WGS analysis are considered as gold standard due to their high discriminatory power when compared to biochemical methods [48]. With ITS sequencing, genetic variations were observed between T. depauwii and T. blankii, with a number of mismatches similar to those of other yeast sibling species [49]. While genetic variation in barcoding genes including ITS is minimal between some species [47], ITS sequencing showed a clear distinction between T. depauwii and the related T. blankii and T. digboiensis, which was confirmed by comparative genomics. Furthermore, all three Tardiomyces species differed in the total number of genes, each exhibiting unique genes that were not found in related species. In addition to genomic differences, phenotypical variation between the three species was observed in enzymatic activity, saline- and osmotolerance. Phenotypical tests, besides the current ITS or WGS analyses, can therefore be used to differentiate T. depauwii from related species in routine clinical settings. It should be noted that few T. depauwii isolates were tested. With the identification of more isolates, the phenotypical variation will likely increase.
In the last decade, the nomenclature of medically important fungi underwent significant changes due to phylogenetic reassignment of genera, leading to name changes of important species including C. glabrata, C. krusei, Candida kefyr, and Candida guilliermondii, renamed as Nakaseomyces glabratus, Pichia kudriavzevii, Kluyveromyces marxianus, and Meyerozyma guilliermondii, respectively [50]. Originally, phenotypic methods were used to characterize yeasts [51]. With phylogenetics, species previously assigned to the genus Candida are often polyphyletic and exhibit widely different characteristics, which do not fit the definition of a genus [52]. The allocation of T. depauwii, T. blankie, and T. digboiensis in a novel genus is more in line with the novel approach [53].
When WGS SNP analysis was applied to the T. depauwii strains, an extremely low genetic diversity of less than 500 SNPs between strains was observed. This, in addition to the estimated TMRCA at 1971, indicates a recent introduction into the human population. The isolation of strains from the oral cavity, sputum and urine suggests that T. depauwii can colonize humans, while its extremely low genetic diversity in combination with its epidemiology suggests a single introduction into the human population and subsequent spread via human-to-human transmission.
Physiological properties
A shared characteristic of species within the Tardiomyces genus is their ability to grow at up to 42 °C, which is rarely observed for other fungi that mostly thrive in the temperature range of 12–30 °C [54]. The genetically related genera Sugiyamaella and Crinitomyces do not grow at 42 °C [55, 56]. The Tardiomyces genus is, therefore, thermotolerant to mammalian temperatures despite all T. depauwii isolates originating from countries with temperate climates. A mammalian reservoir of the latter species could explain such thermotolerance. It is however not likely that T. depauwii is restricted to such high-temperature reservoirs as T. blankii and T. digboiensis strains have also been isolated from varying environments with substantially lower temperatures.
Relatively slow growth was observed for T. depauwii, T. digboiensis, and T. blankii in RPMI 1640 medium with 0.2% glucose when compared to other common yeast species, including C. parapsilosis that is known for its low growth rate [57]. Interestingly, when the experiment was replicated in RPMI 1640 medium with 2% glucose, some T. blankii strains exhibited an elevated growth rate when compared to RPMI 1640 medium with 0.2%. Thus, a lack of glucose is hampering the growth of some T. blankii strains while others seem to be indifferent to its availability. However, the elevated growth rate of these T. blankii strains in this medium was still lower as compared to YPD and SAB broth, indicating multiple nutrients influence the growth rate of these strains [57]. Independently from the available nutrients, T. depauwii and T. digboiensis seem to have an intrinsic low growth speed. As a limitation of this study, it must be noted that genetic diversity of T. depauwii was very low and only a single T. digboiensis isolate was included. Finally, we found that all Tardiomyces strains exhibit the ability to assimilate a wide range of substrates, which was recently also found for T. blankii [58].
Antifungal susceptibility
Due to the slow growth of different Tardiomyces strains, CLSI, EUCAST, and SYO AFST MICs should ideally be read at 48 h, at which antifungals are still potent and MICs can be interpreted reliably [59, 60]. Incubation for longer periods might induce a trailing effect, resulting in unreliable MICs, and is not recommended. Despite a lack of clinical breakpoints and epidemiological cutoff values, T. depauwii can be considered as resistant to fluconazole and voriconazole as high MICs were observed [61]. Susceptibility testing of genetically different strains is required to determine whether this resistance is intrinsic or acquired from a medical or environmental setting [62]. T. blankii and T. digboiensis exhibited elevated MICs for azoles and echinocandins as well, making this likely a worrying genus-wide characteristic.
Despite the apparent low prevalence of T. depauwii and T. blankii invasive infections, the elevated MICs make accurate species identification crucial for effective clinical management. For invasive candidiasis, echinocandins are considered the empirical drug of choice [63]. There are currently no treatment recommendations for this genus, but MICs of micafungin and caspofungin were more favorable than anidulafungin. In one case, echinocandin monotherapy had a favorable outcome [17]. Subsequent step-down azole therapy can be evaluated based on AFST, but may be unfavorable given the high MIC values. Based on MICs reported here, liposomal amphotericin B might represent an appropriate intravenous treatment alternative [64]. A discrepancy was identified between gold-standard AFST (CLSI M27-S4) and SYO, where MICs were elevated using the SYO method. An overestimation of amphotericin B MICs by SYO has also been described for C. auris [65], which can be negated by determining growth inhibition MIC endpoints regardless of the color, as was employed here. A third AFST method using E-test demonstrated MICs after 48 and 72 h were higher than those obtained with regular CLSI AFST but lower than the SYO MICs. The large differences between the echinocandin MICs for C. blankii, e.g., anidulafungin versus micafungin and caspofungin, are remarkable as they have the same target. To our knowledge there are no other species in which such differences have been reported.
In summary, we have described the novel yeast species T. depauwii and propose reassignment of the related C. blankii and C. digboiensis to the novel Tardiomyces genus according to phylogenomic analyses. Shared characteristics of this novel genus are elevated MICs against azoles and some echinocandins, growth up to 42 °C and an overall relative slow growth rate. All characterized T. depauwii strains were of clinical origin with high MICs against fluconazole, voriconazole and anidulafungin, making this a clinically challenging species.
Taxonomy
Tardiomyces B. Spruijtenburg, J.F. Meis & T. de Groot gen. nov.
Tardiomyces (tar.dio.my.ces, from tardio, adjective meaning slow and myces, fungus).
Growth is via multilateral budding of small cells and the formation of pseudohyphae. Species display growth up to 42 °C under standard conditions and generally display a relatively slow growth rate in YPD broth, SAB broth, and RPMI 1640 medium. Fermentation is absent. D-mannose, D-turanose, D-xylose, erythritol, gentobiose, glucoronate, L-arabinose, L-glutamate, L-rhamnose, L-sorbose, xylitol are assimilated and amygladin is not assimilated for all species. Nitrate is not assimilated.
Type species: Tardiomyces depauwii
T. depauwii B. Spruijtenburg, J.F. Meis & T. de Groot sp. nov.
Tardiomyces depauwii (de.pauw.ii, in honor of emeritus prof. Ben de Pauw for his noteworthy contributions to the advancements of clinical mycology).
MycoBank number: 850439. Holotype: CBS 18495, isolated from human blood in Stoke, United Kingdom, preserved in a metabolically inactive state at the CBS culture collection hosted at the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. Living strain ex-type CBS 18495 = NCPF 13064.
On YPD agar after seven days of incubation at 30 °C, colonies are smooth creamy white and soft (Fig. 4A). In YPD broth, budding cells occur singly or in pairs, are ellipsoidal to subspherical, 3–5 µm by 3–7 µm or display formation of pseudohyphae (Fig. 4B). Pseudohyphae bearing branched chains of ovoid cells. Sporulation is not observed in pure cultures on YPD and SAB agar at 30 °C up to ten weeks. Phenotypic tests and growth characteristics are summarized in Table 2. Elevated MICs against multiple azoles and echinocandins were observed according to CLSI microbroth dilution (Table 2).
Tardiomyces blankii (H.R. Buckley & N. van Uden) B. Spruijtenburg, J.F. Meis & T. de Groot comb. nov.
Basionym: Candida blankii H.R. Buckley & N. van Uden. Mycopathologia et mycologia applicata 36(3):257–266 (1968).
Type strain: CBS 1898, isolated from mink organs (Table S1).
Tardiomyces digboiensis (G.S. Prasad, Marilraj, Sood & Lal) B. Spruijtenburg, J.F. Meis & T. de Groot comb. nov.
Basionym: Candida digboiensis G.S. Prasad, Marilraj, Sood & Lal. International journal of systematic and evolutionary microbiology 55(2):967–972 (2005).
Type strain: CBS 9800, isolated from acid tar sludge-contaminated soil (Table S1).
Data availability
No datasets were generated or analysed during the current study.
References
Sharma M, Chakrabarti A. Candidiasis and other emerging yeasts. Curr Fungal Infect Rep. 2023;17:15–24. https://doi.org/10.1007/s12281-023-00455-3.
Kaur H, Shankarnarayana SA, Hallur V, Muralidharan J, Biswal M, Ghosh AK, Ray P, Chakrabarti A, Rudramurthy SM. Prolonged outbreak of Candida krusei candidemia in paediatric ward of Tertiary Care Hospital. Mycopathologia. 2020;185:257–68. https://doi.org/10.1007/s11046-020-00427-y.
Du H, Bing J, Xu X, Zheng Q, Hu T, Hao Y, Li S, Nobile CJ, Zhan P, Huang G. Candida vulturna outbreak caused by cluster of multidrug-resistant strains, China. Emerg Infect Dis. 2023;29:1425–8. https://doi.org/10.3201/eid2907.230254.
Spruijtenburg B, Rudramurthy SM, Meijer EFJ, van Haren MHI, Kaur H, Chakrabarti A, Meis JF, de Groot T. Application of novel short tandem repeat typing for Wickerhamomyces anomalus reveals simultaneous outbreaks within a single hospital. Microorganisms. 2023;11:1525. https://doi.org/10.3390/microorganisms11061525.
Shankarnarayan SA, Rudramurthy SM, Chakrabarti A, Shaw P, Paul S, Sethuraman N, Kaur H, Ghosh AK. Molecular typing and antifungal susceptibility of Candida viswanathii, India. Emerg Infect Dis. 2018;24:1956–8. https://doi.org/10.3201/eid2410.180801.
Lo Cascio G, Vincenzi M, Soldani F, De Carolis E, Maccacaro L, Sorrentino A, Nadali G, Cesaro S, Sommavilla M, Niero V, Naso L, Grancini A, Azzini AM, Sanguinetti M, Tacconelli E, Cornaglia G. Outbreak of Saprochaete clavata sepsis in hematology patients: combined use of MALDI-TOF and sequencing strategy to identify and correlate the episodes. Front Microbiol. 2020;11:84. https://doi.org/10.3389/fmicb.2020.00084.
Yadav A, Jain P, Jain K, Wang Y, Singh A, Singh A, Xu J, Chowdhary A. Genomic analyses of a fungemia outbreak caused by Lodderomyces elongisporus in a neonatal intensive care unit in Delhi, India. MBio. 2023;14: e0063623. https://doi.org/10.1128/mbio.00636-23.
Buckley HR, van Uden N. Five new Candida species. Mycopathol Mycol Appl. 1968;36:257–66. https://doi.org/10.1007/BF02050372.
Pham TT, Dinh KV, Nguyen VD. Biodiversity and enzyme activity of marine fungi with 28 new records from the tropical coastal ecosystems in Vietnam. Mycobiology. 2021;49:559–81. https://doi.org/10.1080/12298093.2021.2008103.
Sandhu DK, Waraich MK. Yeasts associated with pollinating bees and flower nectar. Microb Ecol. 1985;11:51–8. https://doi.org/10.1007/BF02015108.
Yadav A, Jain K, Wang Y, Pawar K, Kaur H, Sharma KK, Tripathy V, Singh A, Xu J, Chowdhary A. Candida auris on apples: diversity and clinical significance. MBio. 2022;13: e0051822. https://doi.org/10.1128/mbio.00518-22.
Arlyapov V, Kamanin S, Ponamoreva O, Reshetilov A. Biosensor analyzer for BOD index express control on the basis of the yeast microorganisms Candida maltosa, Candida blankii, and Debaryomyces hansenii. Enzyme Microb Technol. 2021;50:215–20. https://doi.org/10.1016/j.enzmictec.2012.01.002.
Zaragoza S, Galanternik L, Vazquez M, Teper A, Córdoba S, Finquelievich J. Candida blankii: new agent in cystic fibrosis airways? J Cyst Fibrosis. 2015;14:S140. https://doi.org/10.1016/S1569-1993(15)30492-6.
Al-Haqqan A, Al-Sweih N, Ahmad S, Khan S, Joseph L, Verghese S, Khan Z. Azole-resistant Candida blankii as a newly recognized cause of bloodstream infection. New Microbes New Infect. 2018;26:25–9. https://doi.org/10.1016/j.nmni.2018.06.008.
Kollu VS, Kalagara PK, Islam S, Gupte A. A report of Candida blankii fungemia and possible endocarditis in an immunocompetent individual and the review of literature. Cureus. 2021;13: e14945. https://doi.org/10.7759/cureus.14945.
Mirchin R, Czeresnia JM, Orner EP, Chaturvedi S, Murphy K, Nosanchuk JD. The continuing emergence of Candida blankii as a pathogenic fungus: a new case of fungemia in a patient infected with SARS-CoV-2. J Fungi (Basel). 2022;8:166. https://doi.org/10.3390/jof8020166.
de Almeida JN, Jr CSV, Thomaz DY, Thomaz L, de Almeida RKG, Del Negro GMB, Gimenes VF, Grenfell RC, Motta AL, Rossi F, Benard G. Candida blankii: an emergent opportunistic yeast with reduced susceptibility to antifungals. Emerg Microbes Infect. 2018;7:24. https://doi.org/10.1038/s41426-017-0015-8.
Sathi FA, Aung MS, Paul SK, Nasreen SA, Haque N, Roy S, Ahmed S, Alam MM, Khan S, Rabany MA, Biswas JP, Kobayashi N. Clonal diversity of Candida auris, Candida blankii, and Kodamaea ohmeri isolated from septicemia and otomycosis in Bangladesh as determined by multilocus sequence typing. J Fungi (Basel). 2023;9:658. https://doi.org/10.3390/jof9060658.
Chowdhary A, Stielow JB, Upadhyaya G, Singh PK, Singh A, Meis JF. Candida blankii: an emerging yeast in an outbreak of fungaemia in neonates in Delhi, India. Clin Microbiol Infect. 2020;26:648.e5-648.e8. https://doi.org/10.1016/j.cmi.2020.01.001.
Borman AM, Muller J, Walsh-Quantick J, Szekely A, Patterson Z, Palmer MD, Fraser M, Johnson EM. Fluconazole resistance in isolates of uncommon pathogenic yeast species from the United Kingdom. Antimicrob Agents Chemother. 2019;63:e00211–9. https://doi.org/10.1128/AAC.00211-19.
Sandven P, Bevanger L, Digranes A, Haukland HH, Mannsåker T, Gaustad P, Norwegian Yeast Study Group. Candidemia in Norway (1991 to 2003): results from a nationwide study. J Clin Microbiol. 2006;44:1977–81. https://doi.org/10.1128/JCM.00029-06.
Prasad GS, Mayilraj S, Sood N, Singh V, Biswas K, Lal B. Candida digboiensis sp. nov., a novel anamorphic yeast species from an acidic tar sludge-contaminated oilfield. Int J Syst Evol Microbiol. 2005;55:967–72. https://doi.org/10.1099/ijs.0.63313-0.
Patel MJ, Tipre DR, Dave SR. Isolation and identification of a Candida digboiensis strain from an extreme acid mine drainage of the Lignite Mine, Gujarat. J Basic Microbiol. 2009;49:564–71. https://doi.org/10.1002/jobm.200900084.
Ngom B, Liang Y, Liu Y, Yin H, Liu X. Use of an acidophilic yeast strain to enable the growth of leaching bacteria on solid media. Arch Microbiol. 2015;197:339–46. https://doi.org/10.1007/s00203-014-1051-6.
Arendrup MC, Arikan-Akdagli S, Meinike Jørgensen K, Barac A, Steinmann J, Toscano C, Arsic Arsenijevic V, Sartor A, Lass-Flörl C, Hamprecht A, Matos T, Rogers BRS, Quiles I, Buil J, Özenci V, Krause R, Bassetti M, Loughlin L, Denis B, Grancini A, Lewis White P, Lagrou K, Willinger B, Rautemaa-Richardson R, Hamal P, Ener B, Unalan-Altintop T, Evren E, Hilmioglu-Polat S, Oz Y, Koyuncu Ozyurt O, Aydin F, Růžička F, Meijer EFJ, Gangneux JP, Lockhart DEA, Khanna N, Logan C, Scharmann U, Desoubeaux G, Roilides E, Fe Talento A, van Dijk K, Koehler P, Salmanton-García J, Cornely OA, Hoenigl M. European candidaemia is characterised by notable differential epidemiology and susceptibility pattern; results from the ECMM Candida III study. J Infect. 2023;87:428–37. https://doi.org/10.1016/j.jinf.2023.08.001.
De Carolis E, Vella A, Vaccaro L, Torelli R, Posteraro P, Ricciardi W, Sanguinetti M, Posteraro B. Development and validation of an in-house database for matrix-assisted laser desorption ionization-time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J Clin Microbiol. 2014;52:1453–8. https://doi.org/10.1128/JCM.03355-13.
Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6. https://doi.org/10.1093/bib/bbx108.
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5. https://doi.org/10.1093/nar/gkw256.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6. https://doi.org/10.1093/nar/gkab301.
Community G. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50:W345–51. https://doi.org/10.1093/nar/gkac247.
Kolmogorov M, Yuan J, Lin Y, Pevzner P. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6. https://doi.org/10.1038/s41587-019-0072-8.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. https://doi.org/10.1093/bioinformatics/btp698.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9: e112963. https://doi.org/10.1371/journal.pone.0112963.
Palmer J, Stajich J. Funannotate: a Fungal Genome Annotation and Comparative Genomics Pipeline. v1.5.3. Zenodo; 2019.
Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–8. https://doi.org/10.1101/gr.094607.109.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. https://doi.org/10.1089/cmb.2012.0021.
Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. https://doi.org/10.1186/s13059-019-1832-y.
Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H, Zhang G, Gu YQ, Coleman-Derr D, Xia Q, Wang Y. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–8. https://doi.org/10.1093/nar/gkz333.
Spruijtenburg B, Badali H, Abastabar M, Mirhendi H, Khodavaisy S, Sharifisooraki J, Taghizadeh Armaki M, de Groot T, Meis JF. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg Microbes Infect. 2022;11:2405–11. https://doi.org/10.1080/22221751.2022.2125349.
Spruijtenburg B, Ahmad S, Asadzadeh M, Alfouzan W, Al-Obaid I, Mokaddas E, Meijer EFJ, Meis JF, de Groot T. Whole genome sequencing analysis demonstrates therapy-induced echinocandin resistance in Candida auris isolates. Mycoses. 2023;66:1079–86. https://doi.org/10.1111/myc.13655.
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. https://doi.org/10.1093/ve/vey016.
Ortiz EM. vcf2phylip v2.0: convert a VCF matrix into several matrix formats for phylogenetic analysis. Zenedo; 2019. https://doi.org/10.1038/s41559-019-0982-3.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67:901–4. https://doi.org/10.1093/sysbio/syy032.
Kurtzman CP, Fell JW, Boekhout T, Robert V. Chapter 7 – Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T, Robert V, editors. The yeasts. 5th ed. Amsterdam: Elsevier; 2011. p. 87–110. https://doi.org/10.1016/B978-0-444-52149-1.00007-0.
CLSI. Performance standards for antifungal susceptibility testing of yeasts. 1st ed. CLSI supplement M60. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
Lucking R, Aime MC, Robbertse B, Miller AN, Aoki T, Ariyawansa HA, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, Hawksworth DL, Hyde KD, Irinyi L, Jeewon R, Johnston PR, Kirk PM, Malosso E, May TW, Meyer W, Nilsson HR, Öpik M, Robert V, Stadler M, Thines M, Vu D, Yurkov AM, Zhang N, Schoch CL. Fungal taxonomy and sequence-based nomenclature. Nat Microbiol. 2021;6:540–8. https://doi.org/10.1038/s41564-021-00888-x.
Kim TH, Kweon OH, Kim HR, Lee MK. Identification of uncommon Candida species using commercial identification systems. J Microbiol Biotechnol. 2016;26:2206–13. https://doi.org/10.4014/jmb.1609.09012.
Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gómez-López A, Boekhout T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012;50:3641–51. https://doi.org/10.1128/JCM.02248-12.
Borman AM, Johnson EM. Changes in fungal taxonomy: mycological rationale and clinical implications. Clin Microbiol Rev. 2023;36: e0009922. https://doi.org/10.1128/cmr.00099-22.
Borman AM, Johnson EM. Name changes for fungi of medical importance, 2018 to 2019. J Clin Microbiol. 2021;59:e01811–20. https://doi.org/10.1128/JCM.01811-20.
Daniel HM, Lachance MA, Kurtzman CP. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek. 2014;106:67–84. https://doi.org/10.1007/s10482-014-0170-z.
de Hoog S, Walsh TJ, Ahmed SA, Alastruey-Izquierdo A, Alexander BD, Arendrup MC, Babady E, Bai FY, Balada-Llasat JM, Borman A, Chowdhary A, Clark A, Colgrove RC, Cornely OA, Dingle TC, Dufresne PJ, Fuller J, Gangneux JP, Gibas C, Glasgow H, Gräser Y, Guillot J, Groll AH, Haase G, Hanson K, Harrington A, Hawksworth DL, Hayden RT, Hoenigl M, Hubka V, Johnson K, Kus JV, Li R, Meis JF, Lackner M, Lanternier F, Leal SM Jr, Lee F, Lockhart SR, Luethy P, Martin I, Kwon-Chung KJ, Meyer W, Hong Nguyen M, Ostrosky-Zeichner L, Palavecino E, Pancholi P, Pappas PG, Procop GW, Redhead SA, Rhoads DD, Riedel S, Stevens B, Ota Sullivan K, Vergidis P, Roilides E, Seyedmousavi A, Tao L, Vicente VA, Vitale RG, Wang QM, Wengenack NL, Westblade L, Wiederhold N, White L, Wojewoda CM, Zhang SX. A conceptual framework for nomenclatural stability and validity of medically important fungi: a proposed global consensus guideline for fungal name changes supported by ABP, ASM, CLSI, ECMM, ESCMID-EFISG, EUCAST-AFST, FDLC, IDSA, ISHAM, MMSA, and MSGERC. J Clin Microbiol. 2023;61: e0087323. https://doi.org/10.1128/jcm.00873-23.
Robert V, Cardinali G, Casadevall A. Distribution and impact of yeast thermal tolerance permissive for mammalian infection. BMC Biol. 2015;13:18. https://doi.org/10.1186/s12915-015-0127-3.
Shi CF, Zhang KH, Chai CY, Yan ZL, Hui FL. Diversity of the genus Sugiyamaella and description of two new species from rotting wood in China. MycoKeys. 2021;77:27–39. https://doi.org/10.3897/mycokeys.77.60077.
Sakpuntoon V, Péter G, Groenewald M, Dlauchy D, Limtong S, Srisuk N. Description of Crinitomyces reliqui gen. nov., sp. Nov. and reassignment of Trichosporiella flavificans and Candida ghanaensis to the Genus Crinitomyces. J Fungi. 2022;8:224. https://doi.org/10.3390/jof8030224.
Nordin MAF, Harun WHAW, Razak FA. Antifungal susceptibility and growth inhibitory response of oral Candida species to Brucea javanica Linn. extract. BMC Complement Altern Med. 2013;13:342. https://doi.org/10.1186/1472-6882-13-342.
Opulente DA, Leavitt LaBella A, Harrison MC, Wolters JF, Liu C, Li Y, Kominek J, Steenwyk JL, Stoneman HR, VanDenAvond J, Miller CR, Langdon QK, Silva M, Gonçalves C, Ubbelohde EJ, Li Y, Buh KV, Jarzyna M, Haase MAB, Rosa CA, Čadež N, Libkind D, DeVirgillio JH, Beth Hulfachor A, Kurtzman CP, Paulo Sampaio J, Gonçalves P, Zhou X, Shen XX, Groenewald M, Rokas A, Hittinger CT. Genomic and ecological factors shaping specialism and generalism across an entire subphylum. bioRxiv; 2023. https://doi.org/10.1101/2023.06.19.545611.
Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362:1142–51. https://doi.org/10.1016/S0140-6736(03)14472-8.
Maertens JA. History of the development of azole derivatives. Clin Microbiol Infect. 2004;10:1–10. https://doi.org/10.1111/j.1470-9465.2004.00841.x.
Astvad KMT, Arikan-Akdagli S, Arendrup MC. A pragmatic approach to susceptibility classification of yeasts without EUCAST clinical breakpoints. J Fungi (Basel). 2022;8:141. https://doi.org/10.3390/jof8020141.
Fisher MC, Alasturey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, Bromley M, Brüggemann R, Garber G, Cornely OA, Gurr SJ, Harrison TS, Kuijper E, Rhodes J, Sheppard DC, Warris A, Lewis White P, Xu J, Zwaan B, Verweij PE. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol. 2022;20:557–71. https://doi.org/10.1038/s41579-022-00720-1.
Hoenigl M, Salmanton-García J, Egger M, Gangneux JP, Bicanic T, Araikan-Akdagli S, Alasturey-Ezquierdo A, Klimko N, Barac A, Özenci V, Meijer EFJ, Khanna N, Bassetti M, Rautemaa-Richardson R, Lagrou K, Adam KM, Akalin EH, Akova M, Arsic Arsenijevic V, Aujayeb A, Blennow O, Bretagne S, Danion F, Denis B, de Jonge NA, Desoubeaux G, Drgona L, Erben N, Gori A, García Rodríguez J, Garcia-Vidal C, Roberto Giacobbe D, Goodman AL, Hamal P, Hammarström H, Toscano C, Lanternier F, Lass-Flörl C, Lockhart DEA, Longval T, Loughlin L, Matos T, Mikulska M, Narayanan M, Martín-Pérez S, Prattes J, Rogers B, Rahimli L, Ruiz M, Roilides E, Samarkos M, Scharmann U, Sili U, Resat Sipahi O, Sivakova A, Steinmann J, Trauth J, Ozge T, Van Praet J, Vena A, Lewis White P, Willinger B, Tortorano AM, Arendrup MC, Koehler P, Cornely OA, ECMM Candida III Study Group. Guideline adherence and survival of patients with candidaemia in Europe: results from the ECMM Candida III multinational European observational cohort study. Lancet Infect Dis. 2023;23:751–61. https://doi.org/10.1016/S1473-3099(22)00872-6.
Chen SC, Perfect J, Colombo AL, Cornely OA, Groll AH, Seidel D, Albus K, de Almeida Jr JN, Garcia-Effron G, Gilroy N, Lass-Flörl C, Ostrosky-Zeichner L, Pagano L, Papp T, Rautemaa-Richardson R, Salmanton-García J, Spec A, Steinmann J, Arikan-Akdagli S, Arenz DE, Sprute R, Duran-Graeff L, Freiberger T, Girmenia C, Harris M, Kanj SS, Roudbary M, Lortholary O, Meletiadis J, Segal E, Francisco Tuon F, Wiederhold N, Bicanic T, Chander J, Chen YC, Hsueh PR, Ip M, Munoz P, Spiet I, Temfack E, Thompson L, Tortorano AM, Velegraki A, Govender NP. Global guideline for the diagnosis and management of rare yeast infections: an initiative of the ECMM in cooperation with ISHAM and ASM. Lancet Infect Dis. 2021;21:e375–86. https://doi.org/10.1016/S1473-3099(21)00203-6.
Siopi M, Peroukidou I, Beredaki MI, Spruijtenburg B, de Groot T, Meis JF, Vrioni G, Tsakris A, Pournaras S, Meletiadis J. Overestimation of amphotericin B resistance in Candida auris with Sensititre YeastOne antifungal susceptibility testing: a need for adjustment for correct interpretation. Microbiol Spectr. 2023;11: e0443122. https://doi.org/10.1128/spectrum.04431-22.
Acknowledgements
We would like to thank Dirk Faro for technical assistance.
Funding
This research received support from the Canisius-Wilhelmina Hospital (grant number CWZ_001421). This research was supported by the Brazilian Federal Agency for Support and Evaluation of Graduate Education: National Council for Scientific and Technological Development (CNPq), Brasilia, Brazil (http://cnpq.br/). Bruna Jacomel Favoreto de Souza Lima and Vânia Aparecida Vicente received fellowship from CNPq (grant number 200221/2023-0 and 312811/2018, respectively).
Author information
Authors and Affiliations
Contributions
Conceptualization: Methodology: BS, BJFSL; Validation: BS, BJFSL, AM, SDH; Investigation: BS, BJFSL, STGT, SDH; Resources: AM, CTA, SN, SA, JNAJ, JDN; Data curation: BS, AM, VA, JDN; Original draft preparation: BS, BJFSL; Visualization: BS, BJFSL; Supervision: EFJM, JFM, TDG; Funding acquisition: JFM. All authors reviewed the manuscript
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
15010_2024_2229_MOESM1_ESM.pdf
Supplementary file1 Figure S1 Geographic map depicting T. depauwii isolate overview including clinical information. Figure adapted from //commons.wikimedia.org/wiki/File:North_Sea_location_map.svg and https://en.m.wikipedia.org/wiki/File:USA_Northeastern.png (PDF 216 KB)
15010_2024_2229_MOESM2_ESM.pdf
Supplementary file2 Figure S2 Growth curves of T. depauwii (n = 3), T. blankii (n = 4) and T. digboiensis (n = 1) cultured in RPMI 1640 (0.2% glucose) medium (A), YPD (B) and SAB (C) broth at 35 °C. Strains were grown in eightfold and growth is expressed in average normalized OD values (PDF 129 KB)
15010_2024_2229_MOESM3_ESM.pdf
Supplementary file3 Figure S3 Growth curves of Tardiomyces and common yeast species cultured in RPMI 1640 (2% glucose) medium at 35 °C. On top (A), one strain of each Tardiomyces species is displayed in addition to controls, and on bottom (B) solely Tardiomyces strains are shown. Strains were grown in eightfold and growth is expressed in average normalized OD values (PDF 75 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Spruijtenburg, B., de Souza Lima, B.J.F., Tosar, S.T.G. et al. The yeast genus Tardiomyces gen. nov. with one new species and two new combinations. Infection (2024). https://doi.org/10.1007/s15010-024-02229-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s15010-024-02229-6