Abstract
Purpose
Antimicrobial resistance is a pressing issue in Ukraine, with healthcare-associated infections caused by multidrug-resistant organisms being a major concern. A recent prospective multicenter study revealed a staggering rate of 48.4% antimicrobial resistance to carbapenems among Enterobacterales causing a healthcare-associated infection. We conducted a systematic survey to investigate the incidence rate and incidence density of carbapenemase-producing Gram-negative bacteria (CPGN) among refugees and war-wounded Ukrainians in connection with the German health system.
Methods
From the onset of the war until November 2022, seven Ukrainian patients were admitted to our hospital. Upon admission, screening samples and samples from the focus of suspected infection were taken from all seven patients. The incidence rate and the incidence density of CPGN were calculated as a result of the microbiological findings. We sequenced all CPGN using Illumina technology.
Results
The incidence rate of CPGN at our hospital was 0.06 for 2021 and 0.18 for 2022. All seven Ukrainian patients were infected or colonized with at least one CPGN, including K. pneumoniae (14/25), P. aeruginosa (6/25), A. baumannii (1/25), Providencia stutartii (1/25), C. freundii (1/25), and E. coli (2/25). Genomic surveillance revealed that (i) most frequently detected carbapenemases among all sequenced isolates were blaNDM (17/25) and blaOXA-48 (6/25), (ii) most commonly observed plasmid replicons among the K. pneumoniae isolates recovered from Ukrainian patients were Col(pHAD28) (12/14), IncHI1B(pNDM-MAR) (9/14), IncFIB(pNDM-Mar) (12/14), and (iii) clonal relation between the pathogens of the Ukrainian isolates, but not for the isolates from our hospital surveillance system.
Conclusion
The rising prevalence of community-acquired colonization and infection with CPGN is having a direct effect on the infection prevention measures, such as higher number of isolations, reprocessing of patient rooms, additional microbiological testing and overall organization within hospitals.
Similar content being viewed by others
Introduction
Antimicrobial resistance: political and epidemiological factors get together
Antimicrobial resistance has changed the ways in which therapeutic approaches to bacterial infections and other ailments must be approached. Responsible and judicious use of antibiotics, increased research into developing new and better treatments, and proper education are all essential to mitigating the potentially dangerous consequences of antimicrobial resistance [1]. Antimicrobial resistance is a pressing issue in Ukraine, with healthcare-associated infections caused by multidrug-resistant organisms being a major concern [2, 3]. A recent prospective multicenter study revealed a staggering rate of 48.4% antimicrobial resistance to carbapenems among Enterobacterales causing a healthcare-associated infection [4]. The European Centre for Disease Prevention and Control (ECDC) has warned that the ongoing war in Ukraine could lead to the spread of antimicrobial resistance throughout Europe [5]. Despite this, only a few scientific reports have been published on the matter [6,7,8]. The genomic surveillance of multidrug-resistant organisms from Ukrainian patients in the Netherlands and Germany revealed globally spread epidemic lineages and high number of NDM-1 beta-lactamase. The shortcomings of national surveillance reports are their lack of denominator data. Without denominator data, it is difficult to accurately assess the incidence rate of multidrug-resistant organisms.
We conducted a systematic survey to investigate the incidence of multidrug-resistant Gram-negative bacteria among refugees and war-wounded Ukrainians in connection with the German health system, based on whole-genome sequencing analysis. This study aimed to shed light on the incidence of these bacteria among Ukrainians.
Methods
Recruitment
At our University Hospital, we have 1411 beds across 26 clinics. Every year, we provide care for around 53,600 inpatients and carry out over 274,000 outpatient consultations. As the only university hospital and facility for basic, standard, and maximum health care in the region of Thuringia (a federal state in Germany), our hospital serves over one million citizens. This study encompasses all clinics and institutes of our hospital. From the onset of the war until November 2022, seven Ukrainian patients were admitted to our hospital. Three of these seven patients were hospitalized due to war wounds, while the remaining four were refugees. On-site surgery was performed to treat the wounded soldiers. Upon admission to our clinic, screening samples and samples from the focus of suspected infection were taken from all seven patients. Patient samples were analyzed in an accredited medical microbiology laboratory using standard operating procedures. The incidence rate and incidence density of CPGN were calculated based on the microbiological findings. The incidence rate was calculated for 2021 and 2022 and given in relation to 100 patient cases. The incidence density was also calculated for 2021 and 2022 and given in relation to 1000 patient days.
We sequence all CPGN at our hospital, regardless of whether colonization or infection had occurred. This enabled us to create a database of over a thousand whole-genome sequenced pathogens.
Genomic characterization of multi-drug-resistant isolates via whole-genome sequencing
For microbiological diagnostics, the sample was streaked onto Columbia sheep blood agar (BD, Heidelberg, Germany) and Drigalski lactose agar (Oxoid, ThermoFisher Scientific, Wesel, Germany) for overnight incubation at 37 °C. Colonies were identified by Vitek MS (bioMérieux, Nürtingen, Germany). Antimicrobial susceptibility testing (AST) was performed by determination of minimal inhibitory concentrations (MIC) using Vitek 2 (bioMérieux) and the MICRONAUT-S MDR MRGN Screening microtiter system (Merlin Diagnostika, Bornheim, Germany; distributed by Sifin Diagnostics, Berlin, Germany). Twenty-five isolates from seven Ukrainian patients were sequenced using an Illumina MiniSeq instrument (Illumina, San Diego, CA, USA). DNA was extracted directly from plated microbial cultures using the NexteraTM DNA Flex Microbial Colony Extraction protocol (Illumina, San Diego, CA, USA). The fastq file format was then assembled using Velvet and analyzed with the Ridom SeqSphere + software version 8.4.1. To analyze the species-specific clonal relationship, core genome multilocus sequence typing (cgMLST) was employed. Resistance determinants and plasmids were screened by querying genome assemblies against the ResFinder and PlasmidFinder databases (PlasmidFinder version 2.1.6 (2021-03-27); ResFinder version: 2022-03-17) using the tool ABRicate with minimal coverage and minimal identity settings of 80%, respectively [9,10,11]. The results were then compared to our existing database to assess clonal relatedness and antimicrobial resistance.
Results
War-wounded and refugees within the German health-care system
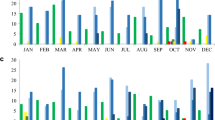
The incidence rate of CPGN at our hospital rose from 0.06 in 2021 to 0.18 in 2022. There was also an increase in incidence density from 2021 (0.08) to 2022 (0.24). This increase was partially attributed to an outbreak of CPGN, which was not associated with Ukrainian patients, resulting in a slight increase in nosocomial cases. However, a significant surge in community-acquired cases was observed in the second and third quarters of 2022 (see Fig. 1), mainly due to the high number of detected CPGN among Ukrainian patients during this period.
All seven Ukrainian patients were infected or colonized with at least one CPGN, including carbapenemase-producing P. aeruginosa, A. baumannii, K. pneumoniae, and Providencia stutartii, C. freundii, and E. coli (see Table 1). The minimum inhibitory concentration values for the antibiotics that were tested have been provided in supplementary information (SI 1). Three wounded soldiers, who had already been treated in Ukraine, were found to have wound infections caused by CPGN, as well as colonization with other carbapenemase-producing species. All patients with an infection received a consultation by an Infectious Diseases physician and were treated with antibiotics tailored to the pathogen detection (e.g., cefiderocol).
High prevalence of antimicrobial resistance genes and mobile genetic elements
The most frequently detected carbapenemases among all sequenced isolates were blaNDM (17/25) and blaOXA-48 (6/25). The ESBL enzyme CTX-M-15 (16/25) and beta-lactamase SHV-182 (8/25) were predominantly found in K. pneumoniae (see Fig. 2A), C. freundii, and E. coli. Notably, the prevalence of these enzymes varied significantly between the species.
Most P. aeruginosa isolates were found to carry blaPAO genes (6/6) as well as blaOXA variants such as blaOXA-395 (3/6), blaOXA-396 (1/6) and blaOXA-488 (2/6) (see Fig. 2B). Notably, one C. freundii isolate was found to contain the newly identified mcr-9 resistance gene [12], which confers resistance to colistin, an antimicrobial drug of last resort used to treat serious Gram-negative bacterial infections. Additionally, several genes encoding resistance to aminoglycosides (aac(6´)-Ib-cr, aph(3´)-VI), chloramphenicol (catA1, catA2), fosfomycin (fosA), fluoroquinolones (oqxA, oqxB, qnrS), sulfonamides (sul1, sul2), and tetracyclines (tet(A)) were observed in the majority of the different species. The resistance genes identified in the Ukrainian isolates are well known and can be found also in our database. There, carbapenemase-forming bacteria were more prevalent in patients with a history of prolonged hospitalization or travel. It is important to highlight that the frequency of detecting resistance genes, particularly carbapenemases, in Ukrainian patients' isolates significantly differs from those in our database. Furthermore, the presence of plasmid replicons was also examined.
The most commonly observed plasmid replicons among the K. pneumoniae isolates recovered from Ukrainian patients were Col(pHAD28) (12/14), IncHI1B(pNDM-MAR) (9/14), and IncFIB(pNDM-Mar) (12/14). Plasmid pNDM-MAR, a member of the IncH plasmid family, is a well-known blaNDM-1-bearing plasmid. In a few of the clinical isolates from our surveillance database, we also detected the IncHI1B(pNDM-MAR) and IncFIB(pNDM-Mar) replicons from K. pneumoniae isolates. The Col(pHAD28) replicon was the most frequently observed and is representative of carbapenemase-producing isolates from our K. pneumoniae database. Plasmid replicons from the same family were also detected in E. coli and C. freundii isolates from Ukrainian patients (see Fig. 3). Furthermore, we have identified the plasmid replicons also in our surveillance database, as illustrated in Fig. 3B.
Comparative analysis of the antimicrobial resistance genes and the plasmid replicons revealed no within-patient plasmid transfer between the different Gram-negative species, causing colonization or infection of the patients.
We investigated the epidemiological connection between Ukrainian isolates and the clinical isolates in our database. We observed a clonal relationship between the pathogens for the Ukrainian isolates, but not for the isolates from our clinical surveillance database. No nosocomial transmissions were detected within our hospital. However, due to the limited medical history and language barrier, it is not possible to evaluate whether there is a possible connection within the home country. For K. pneumoniae, we mostly detected isolates belonging to sequence type 395 (7/13) among the Ukrainian isolates. In our surveillance database, we have identified several MLST sequence types (STs) of bacterial isolates. Among these, ST307 (7% of all K. pneumoniae isolates in our database), ST147 (6%), and ST45 (6%) are the most prevalent in K. pneumoniae. Additionally, we have observed ST395 (6%) to be the most frequently detected ST in our database for this species.
For E. coli, we have sequenced MLST ST131 (E. coli MLST Warwick, 29%) most frequently in our database. Conversely, ST167 and ST46 of the Ukrainian isolates are only sporadically present or not at all in our database. In P. aeruginosa, we have identified MLST ST235 (31%) as the most commonly detected ST in our database. The STs isolated from Ukrainian patients (ST654, ST1047, ST773) have yet to appear in our database. Notably, we isolated pathogens from the same species without high genomic similarity and different antimicrobial resistance profiles within one focus, e.g., wounds (see Table 1, patients #3, #5, #7). This phenomenon is not usually observed for our daily, routinely infectious cases, but from the wounded soldiers.
Discussion
Principal findings
We determined an increase in the incidence rate and incidence density for carbapenemase-producing bacteria from 2021 to 2022 at our hospital. This rise is mainly due to the high number of carbapenem-resistant isolates that we detected in Ukrainian patients. All Ukraine patients, who were admitted at our hospital until November 2022 were colonized or infected with at least one CPGN, which are insensitive to reserve antibiotics. High prevalence of these pathogens causes extensive infection prevention measures. In addition, the risk of nosocomial transmission increases and poses the danger of life-threatening infections that can no longer be treated with antibiotics. Furthermore infections are a major contributor to the rising cost of healthcare [13]. A case–control study revealed that patients who tested positive for carbapenem-resistant Enterobacteriaceae had an elevated risk of having received certain antimicrobials and undergoing invasive procedures. This heightened risk highlights the need for increased education on antimicrobial stewardship and infection control measures to prevent the emergence and spread of carbapenemase-producing bacteria in healthcare settings [14]. Especially the war-wounded patients revealed at least two carbapenemase-forming pathogens per patient, harboring different carbapenemases. This reflects a cross-infection of the wounded patients contaminated with exogenous environmental bacteria and nosocomially transmitted pathogens from first aid on-site as one potential mode of infection and colonization [15]. Alternative explanations may include an origin in the patient’s endogenous colonization flora or transmission events during evacuation transports. A German case report also corresponds to the complexity of caring for war-wounded and other patients from Ukraine and the risk of spreading antimicrobial resistance, but without sequencing results [7].
The increased incidence rate for carbapenemase-producing bacteria within our hospital point up the importance of stringent screening protocols and pre-emptive isolation measures in clinical settings, particularly for war-wounded patients. This proactive approach is critical to ensure the safety of the patients most at risk. Another way to protect vulnerable patient groups from colonization and infection with carbapenemase-producing bacteria is to utilize biocides such as chlorhexidine for bathing patients [16]. However, we have already identified chlorhexidine-resistant K. pneumoniae isolates, raising doubts about the efficacy of this approach [17].
Our report substantiates the recommendations of the ECDC [4], and confirms previously published surveillance data from Germany and the Netherlands, regarding the identified sequence types and resistance genes [6, 8]. In these investigations, blaNDM and blaOXA-48 were also the most commonly detected beta-lactamase. Furthermore, we confirmed the high prevalence for the sequence type ST395 for K. pneumoniae. Regarding the distribution of plasmids, the detected replicons are well-known and have been isolated from patients in West Europe [18]. What is worrying is the prevalence of such plasmids, carrying genes encoding metallo-, extended-spectrum β-lactamase, and a range of resistance genes against other antibiotic groups, even with the small number of cases within our report.
We were able to calculate the incidence rate of carbapenem-resistant strains due to our systematic survey of all patients in our hospital. However, this study is limited to the patients of one hospital. One limitation of the present survey is the small number of patients we could include in our observations. Furthermore, long-read sequencing is the more suitable method to characterize plasmid replicons. For economic reasons, the isolates were not additionally sequenced using a long-read technique.
The incidence rate and incidence density of CPGN have been on the rise, resulting in a greater number of patients being isolated. This has had a significant impact on the organization of examinations and inpatient processes within hospitals, leading to an increase in the number of comprehensive final disinfections. Our clinic has been successful in preventing nosocomial transmission, which validates the efficacy of the hygiene measures and barriers we have implemented. However, the increasing incidence rate of carbapenemase-producing pathogens also highlights the importance of active screening and pre-emptive isolation.
Conclusion
The increasing prevalence of community-acquired colonization and infection with CPGN is increasing epidemiologic pressure on inpatient care. As a result, drastic and costly infection prevention measures are needed to avoid nosocomial transmission and life-threatening infections.
Seamless microbiological surveillance and whole-genome sequencing can create an effective early warning system to combat this issue.
Data availability statement
Whole genome sequencing (WGS) data are available under study ID PRJEB58365 (ERP143427) at the European Nucleotide Archive (ENA).
Abbreviations
- ST:
-
Sequence type
- Cf:
-
C. freundii
- Kp:
-
K. pneumoniae
- Ec:
-
E. coli
- cgMLST:
-
Core genome multilocus sequence typing
- CPGN:
-
Carbapenemase-producing Gram-negative bacteria
- MIC:
-
Minimal inhibitory conentration
- ECDC:
-
European Centre for Disease Prevention and Control
References
Antimicrobial Resistance C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. https://doi.org/10.1016/S0140-6736(21)02724-0. (PMID: 35065702).
Antimicrobial resistance surveillance in Europe 2022 – 2020 data. WHO Regional Office for Europe; 2022; Copenhagen
Kondratiuk V, Jones BT, Kovalchuk V, Kovalenko I, Ganiuk V, Kondratiuk O, et al. Phenotypic and genotypic characterization of antibiotic resistance in military hospital-associated bacteria from war injuries in the Eastern Ukraine conflict between 2014 and 2020. J Hosp Infect. 2021;112:69–76. https://doi.org/10.1016/j.jhin.2021.03.020. (PMID: 33789157).
Salmanov A, Shchehlov D, Svyrydiuk O, Bortnik I, Mamonova M, Korniyenko S, et al. Epidemiology of healthcare-associated infections and mechanisms of antimicrobial resistance of responsible pathogens in Ukraine: a multicentre study. J Hosp Infect. 2022;131:129–38. https://doi.org/10.1016/j.jhin.2022.10.007. (PMID: 36306892).
Operational public health considerations for the prevention and control of infectious diseases in the context of Russia’s aggression towards Ukraine. 2022; Stockholm.
Sandfort M, Hans JB, Fischer MA, Reichert F, Cremanns M, Eisfeld J, et al. Increase in NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae in Germany associated with the war in Ukraine, 2022. Eurosurveillance. 2022;27:2200926. https://doi.org/10.2807/1560-7917.ES.2022.27.50.2200926.
Dietze NTH, Schönherr SG, Lippmann N, Ranft D, Fichtner F, Laudi S, Notov D, Höch A, Kleber C, Lübbert C. Infektionsmedizinische und chirurgische Herausforderungen durch Carbapenem-resistente bakterielle Erreger bei der Versorgung Kriegsverletzter aus der Ukraine. Epid Bull. 2022;36:3–10. https://doi.org/10.25646/10447.
Zwittink RD, Wielders CC, Notermans DW, Verkaik NJ, Schoffelen AF, Witteveen S, et al. Multidrug-resistant organisms in patients from Ukraine in the Netherlands, March to August 2022. Eurosurveillance. 2022;27:2200896. https://doi.org/10.2807/1560-7917.ES.2022.27.50.2200896.
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. https://doi.org/10.1093/jac/dks261. (PMID: 22782487).
Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. https://doi.org/10.1128/AAC.02412-14. (PMID: 24777092).
Ha VTT, Tran LD, Mai NTT, Hirabayashi A, Nguyen ST, Tran HH, et al. Potential spread of mcr-9-carrying IncHI2 plasmids in Enterobacter hormaechei in Vietnam. J Glob Antimicrob Resist. 2021;27:332–4. https://doi.org/10.1016/j.jgar.2021.09.012. (PMID: 34800709).
Stone PW. Economic burden of healthcare-associated infections: an American perspective. Expert Rev Pharmacoecon Outcomes Res. 2009;9:417–22. https://doi.org/10.1586/erp.09.53. (PMID: 19817525).
McAlearney AS, MacEwan SR, Gregory ME, Sova LN, Hebert C, Gaughan AA. Identifying management practices for promoting infection prevention: perspectives on strategic communication. Am J Infect Control. 2022;50:593–7. https://doi.org/10.1016/j.ajic.2021.11.025. (PMID: 34890704).
Asaduzzaman M. Why the war in Ukraine can drive the spread of AMR. In: Europe H, Health Care Without Harm 2022.
Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Heim L, et al. Chlorhexidine versus routine bathing to prevent multidrug-resistant organisms and all-cause bloodstream infections in general medical and surgical units (ABATE Infection trial): a cluster-randomised trial. Lancet. 2019;393:1205–15. https://doi.org/10.1016/S0140-6736(18)32593-5. (PMID: 30850112).
Stein C, Vincze S, Kipp F, Makarewicz O, Al Dahouk S, Pletz MW. Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect Dis. 2019;19:932–3. https://doi.org/10.1016/S1473-3099(19)30427-X. (PMID: 31478517).
Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67:1645–50. https://doi.org/10.1093/jac/dks114. (PMID: 22511638).
Acknowledgements
We are grateful to Monika Cristofolini for her assistance in gathering epidemiological data to determine the incidence rate and incidence density. Her invaluable contribution has been instrumental in helping us to gain a better understanding of the situation.
Funding
Open Access funding enabled and organized by Projekt DEAL. No funding received for this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The first draft of the manuscript was written by CS and MZ. Material preparation, data collection and analysis were performed by CS, RS and MZ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. The authors have no financial or proprietary interests in any material discussed in this article.
Ethical approval
This study was approved by the Ethics committee of the Jena University Hospital (Reg-Nr.: 2023-2918-Daten) conducted in accordance with the relevant ethical guidelines and regulations. Informed consent was waived by the Ethics committee considering the study design and adherence to the relevant guidelines. In particular, study data were deidentified to protect privacy and preserve the confidentiality of the study participants.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stein, C., Zechel, M., Spott, R. et al. Multidrug-resistant isolates from Ukrainian patients in a German health facility: a genomic surveillance study focusing on antimicrobial resistance and bacterial relatedness. Infection 51, 1731–1738 (2023). https://doi.org/10.1007/s15010-023-02061-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-023-02061-4