Abstract
Background and objectives
Diabetes is a chronic disease characterized by high blood sugar. It may cause many complicated disease like stroke, kidney failure, heart attack, etc. About 422 million people were affected by diabetes disease in worldwide in 2014. The figure will be reached 642 million in 2040. The main objective of this study is to develop a machine learning (ML)-based system for predicting diabetic patients.
Materials and methods
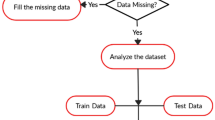
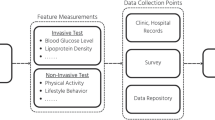
Logistic regression (LR) is used to identify the risk factors for diabetes disease based on p value and odds ratio (OR). We have adopted four classifiers like naïve Bayes (NB), decision tree (DT), Adaboost (AB), and random forest (RF) to predict the diabetic patients. Three types of partition protocols (K2, K5, and K10) have also adopted and repeated these protocols into 20 trails. Performances of these classifiers are evaluated using accuracy (ACC) and area under the curve (AUC).
Results
We have used diabetes dataset, conducted in 2009–2012, derived from the National Health and Nutrition Examination Survey. The dataset consists of 6561 respondents with 657 diabetic and 5904 controls. LR model demonstrates that 7 factors out of 14 as age, education, BMI, systolic BP, diastolic BP, direct cholesterol, and total cholesterol are the risk factors for diabetes. The overall ACC of ML-based system is 90.62%. The combination of LR-based feature selection and RF-based classifier gives 94.25% ACC and 0.95 AUC for K10 protocol.
Conclusion
The combination of LR and RF-based classifier performs better. This combination will be very helpful for predicting diabetic patients.





Similar content being viewed by others
References
American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Supplement 1):S62–9.
Sarwar N, Gao P, Seshasai SR. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease. Lancet. 2010;375(9733):2215–22.
Lonappan A, Bindu G, Thomas V, Jacob J, Rajasekaran C, Mathew KT. Diagnosis of diabetes mellitus using microwaves. J Electromagn Waves Appl. 2007;21(10):1393–401.
Krasteva A, Panov V, Krasteva A, Kisselova A, Krastev Z. Oral cavity and systemic diseases—diabetes mellitus. Biotechnol Biotechnol Equip. 2011;25(1):2183–6.
Nathan DM. Long-term complications of diabetes mellitus. N Engl J Med. 1993;328(23):1676–85.
NCD Risk Factor Collaboration (NCD-RisC). Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 192 million participants. Lancet. 2016;387(10026):1377–96.
Zimmet P, Alberti KG, Magliano DJ, Bennett PH. Diabetes mellitus statistics on prevalence and mortality: facts and fallacies. Nat Rev Endocrinol. 2016;12(10):616.
Bharath C, Saravanan N, Venkatalakshmi S. Assessment of knowledge related to diabetes mellitus among patients attending a dental college in Salem city—a cross sectional study. Braz Dental Sci. 2017;20(3):93–100.
Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, Rao M. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378(9785):31–40.
Iancu, I., Mota, M., & Iancu, E. Method for the analysing of blood glucose dynamics in diabetes mellitus patients. In 2008 IEEE international conference on automation, quality and testing, robotics, vol. 3; 2008. pp. 60–65.
Robertson G, Lehmann ED, Sandham W, Hamilton D. Blood glucose prediction using artificial neural networks trained with the AIDA diabetes simulator: a proof-of-concept pilot study. J Electr Comput Eng. 2012;2011:2–13.
Maniruzzaman M, Kumar N, Abedin MM, Islam MS, Suri HS, El-Baz AS, Suri JS. Comparative approaches for classification of diabetes mellitus data: machine learning paradigm. Comput Methods Programs Biomed. 2017;152:23–34.
Maniruzzaman M, Rahman MJ, Al-MehediHasan M, Suri HS, Abedin MM, El-Baz A, Suri JS. Accurate diabetes risk stratification using machine learning: role of missing value and outliers. J Med Syst. 2018;42(5):92.
Srivastava SK, Singh SK, Suri JS. Healthcare text classification system and its performance evaluation: a source of better intelligence by characterizing healthcare text. J Med Syst. 2018;42(5):97.
Luo G. Automatically explaining machine learning prediction results: a demonstration on type 2 diabetes risk prediction. Health Inf Sci Syst. 2016;4(1):2.
Shakeel PM, Baskar S, Dhulipala VS, Jaber MM. Cloud based framework for diagnosis of diabetes mellitus using K-means clustering. Health Inf Sci Syst. 2018;6(1):16.
Luo G. MLBCD: a machine learning tool for big clinical data. Health Inf Sci Syst. 2015;3(1):3.
Luo G. PredicT-ML: a tool for automating machine learning model building with big clinical data. Health Inf Sci Syst. 2016;4(1):5.
Sahle G. Ethiopic maternal care data mining: discovering the factors that affect postnatal care visit in Ethiopia. Health Inf Sci Syst. 2016;4(1):4.
Shah S, Luo X, Kanakasabai S, Tuason R, Klopper G. Neural networks for mining the associations between diseases and symptoms in clinical notes. Health Inf Sci Syst. 2019;7(1):1.
Bauder RA, Khoshgoftaar TM. The effects of varying class distribution on learner behavior for medicare fraud detection with imbalanced big data. Health Inf Sci Syst. 2018;6(1):9.
Deniz E, Şengür A, Kadiroğlu Z, Guo Y, Bajaj V, Budak Ü. Transfer learning based histopathologic image classification for breast cancer detection. Health Inf Sci Syst. 2018;6(1):18.
Ashour AS, Hawas AR, Guo Y. Comparative study of multiclass classification methods on light microscopic images for hepatic schistosomiasis fibrosis diagnosis. Health Inf Sci Syst. 2018;6(1):7.
Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Laird JR, Suri JS. Wall-based measurement features provides an improved IVUS coronary artery risk assessment when fused with plaque texture-based features during machine learning paradigm. Comput Biol Med. 2017;91:198–212.
Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Suri JS. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J Med Syst. 2017;41(10):152.
Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Khanna N, Suri JS. Calcium detection, its quantification, and grayscale morphology-based risk stratification using machine learning in multimodality big data coronary and carotid scans: a review. Comput Biol Med. 2018;101:184–98.
Bashir S, Qamar U, Khan FH. IntelliHealth: a medical decision support application using a novel weighted multi-layer classifier ensemble framework. J Biomed Inform. 2016;59:185–200.
Zhao X, Zou Q, Liu B, Liu X. Exploratory predicting protein folding model with random forest and hybrid features. Curr Proteomics. 2014;11:289–99.
Sisodia D, Sisodia DS. Prediction of diabetes using classification algorithms. Procedia Comput Sci. 2018;132:1578–85.
Ahuja R, Vivek V, Chandna M, Virmani S, Banga A. Comparative study of various machine learning algorithms for prediction of Insomnia. In: Advanced classification techniques for healthcare analysis; 2019. p. 234–257.
Genuer R, Poggi JM, Tuleau-Malot C. Variable selection using random forests. Pattern Recogn Lett. 2010;31(14):2225–36.
Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2017;20(2):492–503.
Austin PC, Tu JV. Automated variable selection methods for logistic regression produced unstable models for predicting acute myocardial infarction mortality. J Clin Epidemiol. 2004;57(11):1138–46.
Maniruzzaman M, Suri HS, Kumar N, Abedin MM, Rahman MJ, El-Baz A, Suri JS. Risk factors of neonatal mortality and child mortality in Bangladesh. J Glob Health. 2018;8(1):1–16.
Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Programs Biomed. 2017;150:9–22.
Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput Methods Programs Biomed. 2016;126:98–109.
Elssied NOF, Ibrahim O, Osman AH. A Novel feature selection based on one-way ANOVA F-Test for e-mail spam classification. Res J Appl Sci Eng Technol. 2014;7(3):625–38.
Shaharum SM, Sundaraj K, Helmy K. Performance analysis of feature selection method using ANOVA for automatic wheeze detection. Jurnal Teknologi. 2015;77(7):2015.
Wang S, Li D, Song X, Wei Y, Li H. A feature selection method based on improved fisher’s discriminant ratio for text sentiment classification. Expert Syst Appl. 2011;38(7):8696–702.
Cover TM. Geometrical and statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Trans Electron Comput. 1965;14(3):326–34.
Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106.
Hu W, Hu W, Maybank S. Adaboost-based algorithm for network intrusion detection. IEEE Trans Syst Man Cybern B. 2008;38(2):577–83.
Breiman L. Random forest. Mach Learn. 2001;45:5–32.
Liao Z, Ju Y, Zou Q. Prediction of G protein-coupled receptors with SVM-prot features and random forest. Scientifica. 2016;2016:1–10.
Acharya UR, Chua CK, Lim TC, Dorithy, Suri JS. Automatic identification of epileptic EEG signals using nonlinear parameters. J Mech Med Biol. 2009;9(4):539–53.
Ramana BV, Babu MSP, Venkateswarlu NB. A critical comparative study of liver patients from USA and INDIA: an exploratory analysis. Int J Comput Sci Issues. 2012;9(3):506.
Zou Q, Qu K, Luo Y, Yin D, Ju Y, Tang H. Predicting diabetes mellitus with machine learning techniques. Front Genet. 2018;9(515):1–10.
Yu W, Liu T, Valdez R, Gwinn M, Khoury MJ. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med Inform Decis Mak. 2010;10(1):16–23.
Semerdjian J, Frank S. An ensemble classifier for predicting the onset of type II diabetes. arXiv:1708.07480 (2017).
Mohapatra SK, Swain JK, Mohanty MN. Detection of diabetes using multilayer perceptron. In: International conference on intelligent computing and applications, 2019, pp. 109–116.
Pei D, Zhang C, Quan Y, Guo Q. Identification of potential type II diabetes in a chinese population with a sensitive decision tree approach. J Diabetes Res. 2019;2019:1–7.
Acknowledgements
The authors would like to acknowledge the contribution of Statistics Discipline, Science, Engineering and Technology School, Khulna University, Khulna-9208, Bangladesh. The authors also thank to the editor and reviewers for their comments and positive critique.
Funding
No fund received for this project.
Author information
Authors and Affiliations
Contributions
Md. Maniruzzaman: Statistical analysis, draft the original manuscript, and principal investigator and management of the project. Md. Jahanur Rahman: Acquisition of data, interpretation of the results and methodology; Benojir Ahammed: Machine learning concepts and design. Md. Menhazul Abedin: Data preprocessing, English writing, strategy, and interpretation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
No ethics approval is required for this dataset.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1
See Table 9.
Appendix 2
Appendix 3
See Table 13.
Rights and permissions
About this article
Cite this article
Maniruzzaman, M., Rahman, M.J., Ahammed, B. et al. Classification and prediction of diabetes disease using machine learning paradigm. Health Inf Sci Syst 8, 7 (2020). https://doi.org/10.1007/s13755-019-0095-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-019-0095-z