Abstract
Purpose
Significant research has been conducted in the field of brain computer interface (BCI) algorithm development, however, many of the resulting algorithms are both complex, and specific to a particular user as the most successful methodology can vary between individuals and sessions. The objective of this study was to develop a simple yet effective method of feature selection to improve the accuracy of a subject independent BCI algorithm and streamline the process of BCI algorithm development. Over the past several years, several high precision features have been suggested by researchers to classify different motor imagery tasks. This research applies fourteen of these features as a feature pool that can be used as a reference for future researchers. Additionally, we look for the most efficient feature or feature set with four different classifiers that best differentiates several motor imagery tasks. In this work we have successfully employed a feature fusion method to obtain the best sub-set of features. We have proposed a novel computer aided feature selection method to determine the best set of features for distinguishing between motor imagery tasks in lieu of the manual feature selection that has been performed in past studies. The features selected by this method were then fed into a Linear Discriminant Analysis, K-nearest neighbor, decision tree, or support vector machine classifier for classification to determine the overall performance.
Methods
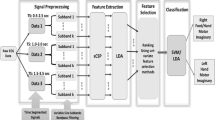
The methods used were a novel performance based additive feature fusion algorithm working in conjunction with machine learning in order to classify the motor imagery signals into particular states. The data used for this study was collected from BCI competition III dataset IVa.
Result
The result of this algorithm was a classification accuracy of 99% for a subject independent algorithm with less computation cost compared to traditional methods, in addition to multiple feature/classifier combinations that outperform current subject independent methods.
Conclusion
The conclusion of this study and its significance is that it developed a viable methodology for simple, efficient feature selection and BCI algorithm development, which leads to an overall increase in algorithm classification accuracy.

Similar content being viewed by others
References
Siuly S, Li Y. Improving the separability of motor imagery EEG signals using a cross correlation-based least square support vector machine for brain–computer interface. IEEE Trans Neural Syst Rehabilit Eng. 2012;20(4):526–38.
Zhou J, et al. Classification of motor imagery EEG using wavelet envelope analysis and LSTM networks. In: 2018 Chinese Control and Decision Conference (CCDC). IEEE, 2018.
Kamousi B, Liu Z, He B. Classification of motor imagery tasks for brain-computer interface applications by means of two equivalent dipoles analysis. IEEE Trans Neural Syst Rehabilit Eng. 2005;13(2):166–71.
Cantillo-Negrete J, et al. An approach to improve the performance of subject-independent BCIs-based on motor imagery allocating subjects by gender. Biomed Eng. 2014;13(1):158.
Vansteensel MJ, Kristo G, Aarnoutse EJ, Ramsey NF. The brain-computer interface researcher’s questionnaire: from research to application. Brain-Comput Interfaces. 2017;4(4):236–47. https://doi.org/10.1080/2326263X.2017.1366237.
Saha S, Mamun KA, Ahmed K, Mostafa R, et al. Progress in brain computer interfaces: challenges and trends. arXiv:1901.03442v1 [cs.HC}, 2019.
Ahn M, Jun SC. Performance variation in motor imagery brain–computer interface: a brief review. J Neurosci Methods. 2015;243:103–10.
Vega R, Sajed T, Mathewson KW, Khare K, et al. Assessment of feature selection and classification methods for recognizing motor imagery tasks from electroencephalographic signals. Artif Intell Res. 2017;6(1):37–51. https://doi.org/10.5430/air.v6n1p37.
Lotte F, Bougrain L, Cichocki A, Clerc M, et al. A review of classification algorithms for EEG-based brain-computer interfaces: a 10 year update. J Neural Eng. 2018. https://doi.org/10.1088/1741-2552/aab2f2.
Shenoy P, Krauledat M, Blankertz B, Rao RPN, Müller KR. Towards adaptive classification for BCI. J Neural Eng. 2006;3:1. https://doi.org/10.1088/1741-2560/3/1/R02.
Jayaram V, et al. Transfer learning in brain-computer interfaces. IEEE Comput Intell Mag. 2016;11:1. https://doi.org/10.1109/MCI.2015.2501545.
Tomioka R, Müller KR. A regularized discriminative framework for EEG analysis with application to brain-computer interface. NeuroImage. 2010;49(1):415–32. https://doi.org/10.1016/j.neuroimage.2009.07.045.
Rahman MKM, Mannan Joadder MA. A review on the components of EEG-based motor imagery classification with quantitative comparison. Appl Theory Comput Technol. 2017;2(2):1–15.
Ghaemi A, et al. Automatic channel selection in EEG signals for classification of left or right hand movement in Brain Computer Interfaces using improved binary gravitation search algorithm. Biomed Signal Process Control. 2017;33:109–18.
Atkinson J, Campos D. Improving BCI-based emotion recognition by combining EEG feature selection and kernel classifiers. Expert Syst Appl. 2016;47:35–41.
Bajaj V, Taran S, Sengur A. Emotion classification using flexible analytic wavelet transform for electroencephalogram signals. Health Inf Sci Syst. 2018;6(1):12.
Taran S, Bajaj V, Siuly S. An optimum allocation sampling based feature extraction scheme for distinguishing seizure and seizure-free EEG signals. Health Inf Sci Syst. 2017;5(1):7.
Wu W, et al. Probabilistic common spatial patterns for multichannel EEG analysis. IEE Trans Pattern Anal Mach Intell. 2015;37(3):639–53. https://doi.org/10.1109/TPAMI.2014.2330598.
Sannelli C, Vidaurre C, Müller KR, Blankertz B. Ensembles of adaptive spatial filters increase BCI performance: an online evaluation. J Neural Eng. 2016;13:4. https://doi.org/10.1088/1741-2560/13/4/046003.
Kevric J, Subasi A. Comparison of signal decomposition methods in classification of EEG signals for motor-imagery BCI system. Biomed Signal Process Control. 2017;31:398–406. https://doi.org/10.1016/j.bspc.2016.09.007.
Blankertz B, et al. The BCI competition 2003: progress and perspectives in detection and discrimination of EEG single trials. IEEE Trans Biomed Eng. 2004;51(6):1044–51.
Siuly S, Li Y, Zhang Y. EEG Signal Analysis and Classification. IEEE Trans Neural Syst Rehabilit Eng. 2016;11:141–4.
Dornhege G, Blankertz B, Curio G, Müller KR. Boosting bit rates in non-invasive EEG single-trial classifications by feature combination and multi-class paradigms. IEEE Trans Biomed Eng. 2004;51(6):993–1002.
Kamousi B, Liu Z, He B. Classification of motor imagery tasks for brain-computer interface applications by means of two equivalent dipoles analysis. IEEE Trans Neural Syst Rehabilit Eng. 2005;13(2):166–71.
Lotte F, Guan C. Regularizing common spatial patterns to improve BCI designs: unified theory and new algorithms. IEEE Trans Biomed Eng. 2011;58(2):355–62.
Shan H, et al. EEG-based motor imagery classification accuracy improves with gradually increased channel number. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE, 2012.
Su Y, Li Y, Wang S. Filter ensemble regularized common spatial pattern for EEG classification. In: Seventh International Conference on Digital Image Processing (ICDIP 2015). Vol 9631. International Society for Optics and Photonics, 2015.
Ramoser H, Muller-Gerking J, Pfurtscheller G. Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans Rehabilit Eng. 2000;8(4):441–6.
Baziyad AG, Djemal R. A study and performance analysis of three paradigms of wavelet coefficients combinations in three-class motor imagery based BCI. In: 2014 5th International Conference on Intelligent Systems, Modelling and Simulation (ISMS), IEEE, 2014.
Dutta S, Chatterjee A, Munshi S. Correlation technique and least square support vector machine combine for frequency domain based ECG beat classification. Med Eng Phys. 2010;32(10):1161–9.
Chandaka S, Chatterjee A, Munshi S. Cross-correlation aided support vector machine classifier for classification of EEG signals. Expert Syst Appl. 2009;36(2):1329–36.
Chandaka S, Amitava C, Sugata M. Support vector machines employing cross-correlation for emotional speech recognition. Measurement. 2009;42(4):611–8.
Krishna DH, Pasha IA, Savithri TS. Classification of EEG motor imagery multi class signals based on cross correlation. Procedia Comput Sci. 2016;85:490–5.
Siuly S, Yan L, Yanchun Z. Cross-correlation aided logistic regression model for the identification of motor imagery EEG signals in BCI applications. EEG signal analysis and classification. Cham: Springer; 2016. p. 153–72.
Zarei R, et al. A PCA aided cross-covariance scheme for discriminative feature extraction from EEG signals. Comput Methods Prog Biomed. 2017;146:47–57.
Yin X, Hadjiloucas S, Zhang Y. Classification of THz pulse signals using two-dimensional cross-correlation feature extraction and non-linear classifiers. Comput Methods Prog Biomed. 2016;127:64–82.
Meng J, et al. Automated selecting subset of channels based on CSP in motor imagery brain-computer interface system. In: 2009 IEEE International Conference on Robotics and Biomimetics (ROBIO) IEEE, 2009.
Reeves RD, et al. Application of correlation analysis for signal-to-noise enhancement in flame spectrometry. Use of correlation in determination of rhodium by atomic fluorescence. Anal Chem. 1973;45(2):253–8.
Taran S, et al. Features based on analytic IMF for classifying motor imagery EEG signals in BCI applications. Measurement. 2018;116:68–76.
Resalat SN, Valiallah S. A study of various feature extraction methods on a motor imagery based brain computer interface system. Basic Clin Neurosci. 2016;7(1):13.
Cantillo-Negrete J, et al. An approach to improve the performance of subject-independent BCIs-based on motor imagery allocating subjects by gender. Biomed Eng. 2014;13(1):158.
Katz MJ. Fractals and the analysis of waveforms. Comput Biol Med. 1988;18(3):145–56.
Higuchi T. Approach to an irregular time series on the basis of the fractal theory. Physica D. 1988;31(2):277–83.
Tolić M, Jović F. Classification of wavelet transformed EEG signals with neural network for imagined mental and motor tasks. Int J Fundam Appl Kinesiol. 2013;45(1):130–8.
Hu J, Xiao D, Mu Z. Application of energy entropy in motor imagery EEG classification. JDCTA. 2009;3(2):83–90.
Abdul-Latif AA, et al. Power changes of EEG signals associated with muscle fatigue: the root mean square analysis of EEG bands. In: Proceedings of the 2004, Intelligent Sensors, Sensor Networks and Information Processing Conference, 2004. IEEE, 2004.
Al-Fahoum AS, Ausilah AA. Methods of EEG signal features extraction using linear analysis in frequency and time-frequency domains. ISRN Neuroscience. 2014;2014:7.
Gurudath N, Riley HB. Drowsy driving detection by EEG analysis using wavelet transform and K-means clustering. Procedia Comput Sci. 2014;34:400–9.
Ho TK. The random subspace method for constructing decision forests. IEEE Trans Pattern Anal Mach Intell. 1998;20(8):832–44.
Mathworks®, MATLAB™ R2017b, Natick, Massachusetts 2017.
Myszewski J, Reina T, Bergendahl E, Rahman M, Development of a classification algorithm for bicep flexion from multi-subject EEG data. In: Proceedings of the Biomedical Engineering Society 2018 Meeting, Oct 2018, Atlanta, Georgia [Online]. Available: http://submissions.mirasmart.com/BMESArchive.Accessed 3 Feb 2019.
Wang H, Zhang Y. Detection of motor imagery EEG signals employing Naïve Bayes based learning process. Measurement. 2016;86:148–58.
Fu R, et al. Improvement motor imagery EEG classification based on regularized linear discriminant analysis. J Med Syst. 2019;43(6):169.
Ye J, et al. Feature reduction via generalized uncorrelated linear discriminant analysis. IEEE Trans Knowl Data Eng. 2006;10:1312–22.
Szuflitowska B, Orłowski P. Comparison of the EEG signal classifiers LDA, NBC and GNBC based on time-frequency features. Pomiary Automatyka Robotyka. 2017;21:39–45.
Bostanov V. BCI competition 2003-data sets Ib and IIb: feature extraction from event-related brain potentials with the continuous wavelet transform and the t-value scalogram. IEEE Trans Biomed Eng. 2004;51(6):1057–106.
Garrett D, et al. Comparison of linear, nonlinear, and feature selection methods for EEG signal classification. IEEE Trans Neural Syst Rehabilit Eng. 2003;11(2):141–4.
Spiewak C, Islam M, Zaman MA, Rahman MH. A comprehensive study on EMG feature extraction and classifiers. Op Acc J Bio Eng App. 2018. https://doi.org/10.32474/OAJBEB.2018.01.000104.
Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat. 1992;46(3):175–85. https://doi.org/10.1080/00031305.1992.10475879.
Prashant G. Decision trees in machine learning. 2017. Available online: https://towardsdatascience.com/decision-trees-in-machine-learning-641b9c4e8052. Accessed 05 May 2019).
Hatamikia S, Nasrabadi AM. Subject independent BCI based on LTCCSP method and GA wrapper optimization. In: IEEE 22nd Iranian Conference on Biomedical Engineering (ICBME), 2015.
Dai M, et al. Transfer kernel common spatial patterns for motor imagery brain-computer interface classification. In: Computational and Mathematical Methods in Medicine 2018 (2018).
Lotte F, Cuntai G, Ang KK. Comparison of designs towards a subject-independent brain-computer interface based on motor imagery. In: 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, 2009.
Acknowledgment
The continued support of the University of Wisconsin-Milwaukee and the University of Wisconsin-Milwaukee College of Engineering and Applied Science, as well as the continued support of our mentors and loved ones throughout our work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Joadder, M.A.M., Myszewski, J.J., Rahman, M.H. et al. A performance based feature selection technique for subject independent MI based BCI. Health Inf Sci Syst 7, 15 (2019). https://doi.org/10.1007/s13755-019-0076-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-019-0076-2