Abstract
Key message
We present a methodological framework that both scientists and supply chain actors can mobilise to organise information at different scales of observation, and further make informed decisions regarding the supply and extraction of bio-molecules from forest biomass. We demonstrate its usefulness for extracting bio-molecules contained in silver fir growing in France.
Context
Numerous bio-active molecules can be extracted from trees at an industrial scale. Supply chain actors play a central role in this emerging bio-economy. However, they do not have enough information and tools to make informed decisions with respect to species, growing locations, or identities of potential suppliers of relevant wood biomass.
Aims
We explore and demonstrate an information chain and methodological framework that can help make three critical decisions regarding the selection of (1) the species containing the desired bio-molecules, (2) the locations where the resource is collected, and (3) the supply chain partners and types of industrial wood by-products necessary to obtain sufficient biomass for industrial extraction.
Methods
The methodological framework provides detailed guidelines and references to select the right combination of sampling protocol, allometric models, chemical analyses, GIS tools, and forest growth and supply chain models in order to produce information for the three decision steps within various regional contexts.
Results
We apply the framework within the context of supply chain actors who are interested in estimating the quantity and diversity of bio-molecules contained in silver fir (Abies alba Mill.) growing in the Grand Est region of France. We show how conflicting environmental, legal and economic constraints can affect the results. We discuss future challenges that need to be tackled to improve the methodological framework.
Conclusion
This study represents a highly detailed overview of the potential bio-molecules contained in a tree species, from its natural habitat or plantation to the end of the regional supply chain. It also represents a step towards the development of a generic knowledge infrastructure and methodology that is necessary to solve various decision-making problems regarding the industrial supply and extraction of high-value bio-molecules.
Similar content being viewed by others
1 Introduction
There is a worldwide interest today in designing a greener economy (D’Amato et al. 2017), in which the potential production of bio-molecules for a diversity of species could be harnessed. By doing so there is some hope that society can substitute petrochemical-based products with renewable ones (Mukhopadhyay 2006; Philp 2018), or design new ones. For instance, bio-molecules extracted from trees (also known as wood extractives) can be used to produce novel and high-value products, e.g. pesticides and fungicides (Anttila et al. 2013; Valette et al. 2017), products for the construction materials industry (Lacoste et al. 2015; Pizzi 2019), food colours and supplements, cosmetics and drugs (Stevanovic et al. 2009; Jablonsky et al. 2017; Bhardwaj et al. 2021).
In practice, supply chain industries from the forestry sector (e.g. sawmills, pulp and paper mills, panel board mills, and lumber brokers) are responsible for finding and extracting bio-molecules from tree species and locations. When doing so, they have to avoid conflicts with the production of other wood products (e.g. wood used for construction, food, industry, energy) and to comply with environmental regulations (e.g. GHG emissions, conservation of forest biodiversity).
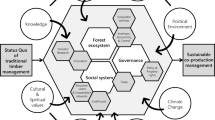
Given the above-mentioned constraints, supply chain actors require prior knowledge about many biological, physical and socio-economic variables at different scales of observation (Fig. 1):
First, supply chain actors require a detailed breakdown of the composition and concentration of bio-molecules (Royer et al. 2013) in order to decide which species to select, trees to fell, wood logs to cut, and biomass components to split, transform and use for industrial extraction (Step 1 in Fig. 1). All trees produce bio-molecules that belong to different groups of polyphenols, including tannins, flavonoids, and lignans. For these bio-molecules, actors need to know how their concentration vary among species (from less than 0.3 to 35% (Willför et al. 2004b; Benković et al. 2014; Kebbi-Benkeder et al. 2015; Bianchi et al. 2016; Kebbi-Benkeder et al. 2017; Nisula 2018)). They also need to know how they vary within a species between the bottom and the top of the trees (Pietarinen et al. 2005, 2008; Kebbi-Benkeder et al. 2017; Nisula 2018; Brennan et al. 2020a, 2020b, 2020c), between young and old trees (Dedrie et al. 2015; Jyske et al. 2014), and between the biomass components of trees (heartwood, sapwood, bark, knots and branches, and branch insertions) (Sinclair and Dymond 1973; Kebbi-Benkeder et al. 2015). For example, knots contain 5–20% polyphenols, like lignans in conifers and flavonoids in deciduous species (Holmbom et al. 2003; Willför et al. 2003, 2004a, 2004b, 2004c, 2005; Pietarinen et al. 2005; Sekine et al. 2013; Kebbi-Benkeder et al. 2015; Nisula 2018). Likewise, they need to know whether the diversity and concentration of bio-molecules differ from the normal level. This may occur in response to some environmental factors at the growth site (Kilulya et al. 2014; Piispanen et al. 2008), or around scars and specific biomass components after trees have been pruned, de-barked or wounded due to wind breakage of branches, silviculture or herbivory (Lewinsohn et al. 1991; Kopaczyk et al. 2020).
Then, supply chain actors need to locate forest catchments that contain the requested quantities of bio-molecules (Step 2 in Fig. 1). They need to do that to identify the land owners they would contract with for using already felled trees or harvesting new ones (IGN 2018). But getting this information requires an extended knowledge of how the composition and dynamics of the bio-molecules at the scale of a population of standing and felled trees is impacted by various biological and environmental disturbances, as well as by silvicultural practices. Eventually they also need to know whether there is a suitable road infrastructure, and whether there are nearby industries that procure wood from the forest landowner. Geographic information systems (GIS) could be used to synthesise and project across space and time the composition and quantities of bio-molecules for any given species. But to the best of our knowledge, there is no published work or map of this kind, not even for the most advanced studies in the fields, such as in Canada (see Royer et al. 2010, 2012, 2013) or in Scandinavia (Holmbom 2011; Näyhä et al. 2011; Bergström and Matisons 2014; Routa et al. 2017; Nisula 2018; Verkasalo et al. 2021).
Next, supply chain actors need to select suppliers that can transform wood biomass into extractable wood fractions (Step 3 in Fig. 1). The problem is that detailed knowledge of supply chains is rarely shared. They represent complex and strategic networks of forest industries, involving multiple pathways and loops of purchasing, selling, and the transformation of unprocessed and processed wood biomass (Velema 2004; Roitto et al. 2016; Routa et al. 2017; Pichancourt et al. 2018; Hassegawa et al. 2018). The lack of such public data makes it difficult to predict where and how much of the richest fraction of bio-molecules of the biomass components are separated during biomass processing (e.g. bark, sawdust, chips and knotwood), where they are aggregated into composite by-products that could be more cost-effectively bought and collected for the industrial extraction of bio-molecules (Holmbom et al. 2003; Nabarlatz et al. 2007; Rustad et al. 2011) and, finally, where they could be collected without disturbing traditional uses of forest biomass (Velema 2004; Roitto et al. 2016; Pichancourt et al. 2018; Hassegawa et al. 2018; Verkasalo et al. 2021).
Finally, supply chain actors need to select an industrial extractor specialised in biorefining (Step 4 in Fig. 1). However, beyond the cost of industrial extraction itself, they need to know whether the process of extraction actually improves the carbon balance of their activity, as opposed to if they were selling their wood biomass for another use (e.g. for bio-energy). By estimating that, they may be able to claim (additional) carbon credits (Brunet-Navarro et al. 2016; Pichancourt et al. 2018).
Altogether, the strategic pieces of information that we present at the different scales of observation form what is known as a “knowledge infrastructure”. Without it, supply chain actors may not have the necessary “visibility horizon” (as per Carter et al. 2015) to adapt their activities to this new bio-economy. This knowledge infrastructure is not easy to produce and mobilise. In this study, we show how it can be achieved by developing a generic methodological framework that can be used to select the right data, models and analyses to produce information at different scales (Fig. 2). We then apply it to a case study where we worked with supply chain actors from the Grand Est region of France who are interested in the industrial extraction of bio-molecules contained in silver fir (Abies alba).
2 Methodological framework
2.1 Selection of the best tree species and biomass components to be used
For this first step (Step 1 in Fig. 1), the objective is to estimate the quantity of bio-molecule i that can be extracted from biomass components j (e.g. stem, bark, knots, branch insertions, branches), at tree height k for a particular tree species. To achieve this, we need to independently estimate the volume (Vjk [m3]) and the basic wood density (WDjk [kg.m− 3]) for each biomass component, as well as the dry mass proportion of the extractives (ECijk [kg.kg− 1] or [%]). Then, using simple arithmetic, we can deduce the total quantity of the extractable bio-molecule (Bijk [kg]), such that:
To carry out this simple calculation, four prior scientific tasks need to be performed:
2.1.1 Selection, measurement and harvesting of trees
Studying the variability of bio-molecules at both a tree level and a species level is expensive and time-consuming. Therefore, the sampling effort at both levels should be adapted to the objective, time and money (see, e.g., Royer et al. 2010, 2012, 2013, Kebbi-Benkeder et al. 2017; Billard et al. 2020, Brennan et al. 2020a, 2020b or Brennan et al. 2020c). Regardless of the sampling effort, the spatial location and biological characteristics of standing trees must be measured for further allometric and chemistry analyses (e.g. breast height diameter (dbh), tree height, crown height, height of the first living branch, stem and branch diameters).
2.1.2 Preparation for allometric and chemical analyses
This task depends on the tree phenotype and sampling effort. When many species need to be extensively compared, then 1–2 transverse discs of several centimetres are sufficient, sawn at the stump and at the height of 1.3 m from the base (Kebbi-Benkeder et al. 2015) or, if possible, at 5 m if the species has butt swelling (like for some tropical trees) (Santana and Okino 2007). If more intensive analyses are needed to study the differences in bio-molecules between the biomass components, several evenly spaced discs are cut (e.g. 2–15), and their position along the trunk is recorded (Kebbi-Benkeder et al. 2017). If data must be further segregated based on how biomass components are used (e.g. for industrial uses, construction or wood for energy), we suggest sawing the discs along the trunk, from the base to the upper limit of wood suitable for industrial uses (Billard et al. 2020; Brennan et al. 2020a, 2020b, 2020c).
2.1.3 Estimation of the basic density and relative volume of biomass components
This task can be skipped if prior measurements of biomass have been performed directly on standing trees, or indirectly using allometric equations, i.e. predicted from stem or branch diameters (as described in detail by Gómez-García et al. (2015)). However, recent studies suggest that current manual and allometric knowledge may be misleading and should be improved (Bauer et al. 2019; Billard et al. 2020). In this case, some of the discs (after being oven-dried) should be analysed with an X-ray computed tomography scanner (CT-scanner) (Longuetaud et al. 2016; Billard et al. 2020)) to estimate the precise volume and basic density [kg.m− 3] of all of the biomass components. The statistical method, assumptions and source code to achieve this are described in details in Longuetaud et al. (2016).
2.1.4 Estimation of the composition and concentrations of bio-molecules
Discs and knots are then independently crushed to extract the bio-molecules of interest, as described, e.g., in Brennan et al. (2020a, 2020b, 2020c). The lab process should match the actual industrial process of extraction used by biorefineries and legal environmental constraints. Briefly, discs are milled, dried, and processed with a solvent in an accelerator for extraction, before estimation of the dry yield. Various solvents, such as water, ethanol, a mixture of the two, or even more efficient ones such as the toluene/ethanol mixture (if authorised by the legislation). Liquid chromatography is then followed by mass spectroscopy (LC-MS) and nuclear magnetic resonance spectroscopy (13C NMR) to quantify and identify polyphenolics and others.
Once the distribution model of extractives has been inferred for the desired species, it may be used together with other methods to support decision-making. However, the first step can still be repeated again for two independent reasons: either when it is a question of geographic region where the species ecology is known to be different, or when better pre-screening of some of the tree biomass components is necessary. This can be the case when the harvesting of wood biomass is legally constrained by prioritisation rules in some countries. For this purpose, we will show how it is possible to restructure the dataset in the application example section.
2.2 Selection of the best geographical location for harvesting
The methodological objective of Step 2 (Fig. 1) can be broken down into two tasks:
2.2.1 Scaling up the information about bio-molecules in the standing forest resource
The common method here is to develop a GIS map containing information about the composition and quantity of bio-molecules in the standing forest resource and felled logs. These layers of information can be crossed with other information layers, e.g. with the road network, the land register, or the distance from the nearest forest industries. This task obviously depends on the availability of spatial data related to the forest resource. The latter can be obtained from any National Forest Inventory (NFI) programme. NFIs provide statistically representative information (e.g. forest composition and structure, dendrometric data) on a large spatial scale based on sampling methods, geo-statistical interpolation and allometric models (Vidal et al. 2016). Various statistical methods can be used to represent NFI data on a GIS map, depending on the quality of the dataset (Vidal et al. 2016; Köhl et al. 2006). Equation 1 can then be combined with NFI data in a GIS to estimate the quantity of extractives.
2.2.2 Projecting standing resource information over time for different socio-ecological scenarios
The main method consists in designing and using a forest growth model to project the biomass and the composition in bio-molecules for different tree size classes and harvesting scenarios (Wernsdörfer et al. 2012). For this task, several types of forest growth models can been used Weiskittel et al. (2011). Mäkelä and Valentine (2020) provide guidelines to select the right model, depending on the objective, scale of projection, level of forest complexity, data available, and computational limitations. Once the right model is selected, we can produce GIS layers to project the spatial forest distribution and biomass density of bio-molecules in both standing trees and felled logs for different time horizons (Colin et al. 2017).
2.3 Selection of the best suppliers to buy by-products for extraction
The methodological objective of Step 3 (Fig. 1) can be broken down into two tasks:
2.3.1 Reconciling supply chain data
Since private supply chain data are strategic and frequently not shared, public data can be used as a surrogate. They usually present aggregated quantities of raw material that are harvested, and the annual proportions that are processed at a regional or a national scale by different groups of industries, e.g. all sawmills (Martin 2009; Lenglet et al. 2017; Pichancourt et al. 2018). However, this type of supply chain data is frequently heterogeneous, since it comes from different sources and scales of observation, and may thus benefit from being reconciled using optimal material flow analyses (Cencic 2016; Lenglet et al. 2017).
2.3.2 Connecting resource growth dynamics and supply chain data
The supply chain model is then used as a data integration tool to represent the supply chain structure and the associated flow of wood material and bio-molecules (Holmbom 2011; Bergström and Matisons 2014; Routa et al. 2017; Pichancourt et al. 2018). It is then used to test alternative scenarios of re-orienting wood biomass for extraction in order to find the right supplier and to quantify the sensitivity of the flow of bio-molecules in response to various levels of social and environmental risks and uncertainties (Vieira et al. 2020). There are different types of supply chain models and not all can achieve what has been mentioned so far. The reader may refer to Min and Zhou (2002), Vahid et al. (2016), and Pichancourt et al. (2018) or Vieira et al. (2020) among others, to select the most appropriate model.
2.4 The regional impact of redirecting wood biomass for industrial extraction
Finally, the methodological objective of the last (Step 4 in Fig. 1) is to assess the regional impact of constraints of re-routing wood biomass for other end-uses, such as industrial extraction. For instance, analyses of GHG emissions are considered a common business practice that requires standard calculations defined by the IPCC (Lindner et al. 2012; Pichancourt et al. 2018). A list of the supply chain models that include these calculations, with their pros and cons, can be found in Brunet-Navarro et al. (2016) and Pichancourt et al. (2018).
3 Application example: silver fir in the Grand Est region (North-Eastern France)
Our aim here was to show how this methodological framework could be applied, to help supply chain actors from the Grand Est region to extract high-value bio-molecules contained in the common tree species silver fir (Abies alba Mill.), using an industrial scale biorefinery. The application example combined many of the classic supply chain management problems predicted by the related theory (Carter et al. 2015), i.e. poor awareness and poor data sharing and coordination practices regarding the structure and dynamics of the entire supply of forest products (Ficko et al. 2011; Pichancourt et al. 2018; ADEME 2019). The tailoring of methods that we produced represents three years of multidisciplinary scientific work, plus feedback from various forest supply chain actors that we met in the region, during multiple thematic meetings, workshops and formal interviews.
3.1 Biomass components useful for extracting bio-molecules
We first estimated the total quantity of bio-molecules contained in most biomass components of eight silver fir trees harvested in 2018 at 49.97∘N, 4.06∘ E. The harvest took place in a 40-year-old naturally regenerated mixed-forest stand with 90% silver fir, 10% Norway spruce, and a small number of miscellaneous broad-leaved species. The wood chemistry analysis showed that the composition of bio-molecules per unit of wood biomass [kg/kg] varied between the biomass components. Stemwood represented the largest fraction of bio-molecules, followed by bark and knotwood with much smaller fractions of the total tree biomass.
CT-scan analyses showed that the variation in the total biomass of bio-molecules was a consequence of differences in the relative volume, wood basic density, and extractive concentration between the biomass components (Fig. 3). Dry weight of bio-molecules per cubic metre of stem volume was estimated to be \(\sim 15~kg.m^{-3}\), independent of tree size, and \(\sim 12~kg.m^{-3}\) and \(\sim 1~kg.m^{-3}\) in bark and knotwood, respectively. The highest concentrations of bio-molecules on a dry weight basis were observed in bark (0.2–0.25 kg.kg− 1) and knotwood (0.1–0.2 kg.kg− 1), and the lowest concentrations in stemwood (< 0.05 kg.kg− 1). We could not produce any information about bio-molecules for branches. Given that knots represent only \(\sim ~\nicefrac {1}{100}^{th}\) of the total above-ground volume of silver fir, their bio-molecules was estimated to be no more than \(\sim ~\nicefrac {1}{500}^{th}\) of the total biomass of a tree, even if knots were the densest wood component (700 ± 41 kg.m− 3), and almost twice as dense as stemwood (374 ± 18 kg.m− 3). Similarly, bio-molecules in bark represented no more than \(\sim ~\nicefrac {1}{50}^{th}\) of the total biomass of silver fir, even if barkwood density (514 ± 19 kg.m− 3) represented \(\sim ~\nicefrac {4}{3}^{rd}\) of stemwood density.
Bio-molecules that can be extracted from five biomass components of silver fir: (1) bark, (2) stem, (3) knot, (4) insertion of the branch, (5) branch (\(\varnothing \) = [0–4 cm, 4–7 cm, > 7 cm]). We measured the basic wood density (WD: [kg.m− 3]), the relative volume (RV: [%] of the above-ground volume of the tree with the exclusion of leaves), and the graphs represent the concentration in extractives, EC [kg.kg− 1]. For every biomass component, potential conflicts (construction and industrial use in this case) are presented if the wood materials are used for molecular extraction on the same harvesting sites
The concentration of bio-molecules varied along the tree. However, given the national priority rules for using wood biomass in France (ADEME (2019, p. 8): construction wood & industrial wood products > wood for chemical industry > energy wood), we categorised the results into industrial wood and energy wood (dbh ≤ 20 cm) and into sawlogs (dbh > 20 cm). Particularly higher grade bottom logs of silver fir are usually prioritised by sawmills for the supply of construction wood or packaging. Upper logs are prioritised for industrial wood (e.g. for panel or pulp&paper making), either coming from the stem part above the lowest living branches, or from young trees removed from thinning. Sawdust, knots and bark by-produced by sawmills, pulp and paper mills are usually sold there for making either panels, but mostly for energy wood in the form of pellets or cogeneration. Given those constraints, we show that knotwood, which are easier to obtain, contains more extractives than tree segments that are used for sawn timber (\(\sim \) 18%) or tree parts that are used for industry or energy wood (\(\sim \) 10%).
Finally more detailed wood chemistry analyses revealed that bark and knotwood samples both contained a large diversity of families of extractable bio-molecules (Table 1). Bark contained 13 bio-molecules (nine identifiable ones) from different families, mostly flavan-3-ol, phenolic compounds and tannins. Knots contained seven bio-molecules at 10 to 100 times lower concentrations, from the following families of molecules: lignans, phenolics, terpene and from unknown origins. The largest diversity of bio-molecules was found in knotwood. Many of these bio-molecules are biologically active and have nutraceutical properties that can be used in food supplements (Bhardwaj et al. 2021). Moreover, isorhamnetin flavonoids found in barkwood are known for their anti-inflammatory and anti-tumoral effects, and lignans from knotwood for its anti-tumoural properties (Gong et al. 2020; Bhardwaj et al. 2021).
3.2 Locations where trees contain the desired bio-molecules
Combining GIS projections and French NFI data, we projected the regional distribution and variability of the biomass of a diversity of bio-molecules contained in one hectare of forest with silver fir trees (Fig. 4). As an example knotwood of standing trees in one hectare of forest from the Vosges mountains should contain \(\sim \) 1 to 40 kg of different types of lignans with potentially useful nutraceutical properties; whereas two to six hectares would be necessary to obtain the same amount of isorhamnetin from barkwood (known for its anti-tumoural properties).
Locations and amount of bio-molecules per hectare of standing forest containing silver fir in the Grand Est region. The results were generated using the GIS platform that can be found at http://extraforest.ign.fr/
We then combined the GIS information with a size structured forest growth model, calibrated on the basis of regional inventory data (Wernsdörfer et al. 2012; Colin et al. 2017) (Table 2). We used it to project the expected space, time and tree size structure of the bio-molecules for the next 20 years, in both standing trees and trees that were expected to be logged for other uses (i.e. for construction, industrial and energy wood). In our case, the future scenarios of potential harvesting were constructed in collaboration with relevant forest agencies. We estimated the future biomass of bio-molecules in standing and felled logs, and the uncertainty was estimated using a bootstrapped sensitivity analysis (1000×). These results are not presented in this section, but were used to produce molecular biomass information along the supply chain in the following section.
3.3 Bio-molecules contained in by-products coming from suppliers
We used the 2017–2050 projections for the biomass of bio- molecules contained in felled logs as input for a supply chain model (Pichancourt et al. 2018, 2020). We projected them along a scenario of a regional supply chain reconstructed using several sources of data (Fig. 5): (i) the latest 2017 data for the Grand Est region from the French national forest and timber supply chain survey (Normandin 1990; Paquet and Deroubaix 2003; AGRESTE 2017); (ii) results from a previous material flow analysis for the region (Lenglet et al. 2017; Pichancourt et al. 2018); and (iii) data that we collected during semi-structured interviews with specific industrial actors, under confidential data agreements.
The silver fir supply chain operating in the French Grand Est region was modelled using the flux manager module of a wood supply chain analysis software (Pichancourt et al. 2018). In this scenario, 20% of the by-products (bark, knots) of the manufacturing of sawn timber, pulp and paper (initially sold for energy wood) were reallocated to extract bio-molecules.
For the scenario, 1000 runs of pair-wise Monte Carlo uncertainty and sensitivity analyses applied on various parameters of the supply chain model showed that significant annual quantities of the previously mentioned bio-molecules could be extracted during the next 20 years from the by-products made of knotwood (Fig. 6a) or barkwood (Fig. 6b) from sawmills and paper mills.
Bio-molecules contained in wood by-products from sawmills and paper mills. Predicted cumulated quantities (2017–2037) of families of bio-molecules that can be extracted from (a) knotwood by-products and (b) barkwood by-products, produced by forestry industries using silver fir harvested across the 1,465,034.8 ha of forests present in the Grand Est region of France. Values were estimated using the wood supply chain software presented in Pichancourt et al. (2018) and connected with a bio-chemistry module (Pichancourt 2020). Bars represent the 95% confidence intervals estimated using 1000x bootstrapping
3.4 Regional impact assessment of the extraction of bio-molecules
Reorienting part of the by-products for industrial extraction of bio-molecules is expected to represent a very small fraction of the wood biomass (measured in product biomass [kg]) that should have been used for energy for the next 20 years (Fig. 7a). This amount is also insignificant when compared to the total wood biomass fraction used to build all other classes of end-products (buildings, furniture, paper, wrapping). For carbon balance accountability, the production and use of these bio-molecules is predicted to reduce the carbon footprint of the regional supply chain (re-scaled as CO2 content [kg CO2eq]: Fig. 7b) by avoiding the burning and release into the atmosphere of several thousand tonnes of CO2 from wood residuals over the next 20 years.
(a) Expected cumulated biomass of the various types of wood products, including the bio-molecules (extractives), that can be produced by 2037 in the Grand Est region of France. (b) Cumulated GHG emissions saved by re-routing by-products produced by the forestry industries, for the industrial extraction of bio-molecules. Values were first estimated per hectare using the carbon accounting module of a wood supply chain software presented in Pichancourt et al. (2018). They were then re-scaled for the 1,465,034.8 ha of forests containing silver fir present in the Grand Est region. The error bars in (a) and grey ribbon in (b) represent 95% confidence intervals using 1000x bootstrapping
4 Discussion
In our study, we demonstrated the possibility to produce and articulate diverse information about bio-molecules obtained at different scales. Based on this unique experience, we now discuss some problems and methodological challenges that can emerge for this procedure.
4.1 Challenges for the selection of the best species and biomass components
The supply chain actors that we met had quantitative knowledge of the extractable fraction of lignocellulosic macro-molecules, because of their historical economic interest in producing wood products and biofuel, their low requirements for homogeneity, and low variability among species (Naik et al. 2010; Jayasinghe and Hawboldt 2012). However, their knowledge of other bio-molecules was not as good for the reasons we mentioned above. In this first exploratory study, we show that intensive data collection and analysis at a tree level were required to provide a complete picture of the bio-molecules for silver fir, justifying the fact that we only processed eight trees. The same problem has been encountered for similar initiatives in Canada (Royer et al. 2010; Royer et al. 2012; Royer et al. 2013). Before extending our work to other species, our results would have to be validated on more trees and compared to direct allometric model approaches that predict the biomass of some tree components (Gómez-García et al. 2015). Our allometric models could further be extended to predict how the biological and chemical characteristics of the biomass components vary with local ecological factors (Piispanen et al. 2008; Rivoire et al. 2010; Genet et al. 2011; Hounzandji et al. 2014; Kilulya et al. 2014), or with other environmental stressors on standing trees (Lewinsohn et al. 1991; Kopaczyk et al. 2020), or even on felled trees at logging or stocking sites (Anttila et al. 2013). However, regardless of how well we can simplify the sampling protocol, the choice of one species or biomass component is not only about wood chemistry. Supply chain actors that we met mentioned the need to consider the monetary value of bio-molecules and market demand (Fig. 3). They want to run cost-benefit analyses, rank the species and biomass components, and decide whether it is worth moving away from other timber uses. In this case, multi-criteria optimisation methods need to be developed.
4.2 Challenges for the selection of the best harvesting locations
To the best of our knowledge our work represents the first attempt to publish a distribution map or temporal projections of the composition and quantity of bio-molecules per hectare for a species (see Fig. 4). Nevertheless, several methodological challenges still remain to be solved.
The first challenge is related to the resolution of the NFI sampling scheme. The number of trees currently sampled statistically limits the spatial scale at which we can produce results for bio-molecules across space and time without producing interpolation errors (IGN 2018). Because the number of silver fir trees varied from one place to another, it was difficult to predict the minimal scale at which we can make interpolation beforehand. The GIS platform that we developed automatically integrates these constraint, prevents further analyses, and suggests that the user should select a larger area. However, our interactions with forest actors during thematic meetings and workshops suggested that the scale at which they could make predictions for silver fir was not always adequate. For instance, small sawmills preferred to obtain information about bio-molecules at the scale of a small harvesting catchment or even of a single private forest farm, which we could not provide. For them, we would need to develop better methods to combine data from current NFI sampling schemes with higher resolution of remote or local inventory data (see, e.g., Kangas et al. 2019).
The second challenge is related to determining the best forest resource catchments to invest in for future industrial extraction. In our case, criteria other than biological ones would need to be considered, such as the cost of transportation, the presence/absence of a road network, of forestry infrastructures, of land conflicts between timber objectives and some ecosystem services (e.g. carbon sequestration, biodiversity conservation, cultural services). Our GIS platform already had these layers of information, but the supply chain actors that we met were eager to select the best location by considering this diversity of social-ecological criteria. We think that the spatial optimisation algorithms presented by Ananda and Herath (2009) could be of use to solve these multi-dimensional problems.
The last challenge is related to the use of GIS platforms for the prospection of bio-molecules at a large scale. Our platform was tested on the Grand Est region for precise molecular estimates, but could be used at a national scale for less precise metrics. Moreover, some scientists and supply chain actors from neighbouring countries asked us to extend this study to better match the spatial structure of forest resource catchments and some supply chains (Mukhopadhyay 2006). However, the problem is that NFIs are frequently not harmonised between neighbouring countries (Vidal et al. 2016), and even if data harmonisation and infrastructure are operational (as planned between the EU member states), this may not be sufficient to implement a cross-country forest information system for strategic reasons (Baycheva-Merger and Sotirov 2020). Targeted R&D initiatives like ours may help solve this problem.
4.3 Challenges for the selection of the best suppliers
For the supply chain step, we demonstrated that it was possible to use models to predict the best supplier and the biomass and diversity of bio-molecules that could be extracted. As of that point, we scan anticipate three remaining challenges.
First in our supply chain projections, we included various sources of uncertainties (see Figs. 3 and 4). However, others have not yet been factored in, including those found in the literature. A first source of uncertainty is related to knot removal by transformation industries. Interviews that we conducted with industrial actors showed that sawmills and paper mills use machines that cut a bit more wood around the knots (1–3%) than what was measured by CT-scan (\(\thicksim ~1\%\)). The exact quantity can change from one mill to another, depending on the exact parameterisation of the machine used. However, we do not know its variability at a regional scale. A second source of uncertainty is related to carbon balance calculations when modelling the industrial process of extraction. We do not have any information about the carbon that would be emitted during the process. However, various industrial extraction pathways can be used, each with its own GHG emission efficiency (Liao et al. 2020). Including these two sources of uncertainties into supply chain projections may change the final results we presented in Figs. 6 and 7.
Next, using our simulation-based supply chain model, we analysed whether re-routing the biomass from two supply chain actors could significantly increase the supply of bio-molecules. The non-analytical nature of our model required us to make targeted sensitivity analyses by presenting a specific scenario of supply chain adaptation. However, extending this logic to analyse the sensitivity of the entire supply chain structure in relation to each and every industry would have been preferable. Global sensitivity analyses of this kind are complex analytical questions, and the literature on the topic is sparse (Rabelo et al. 2011). An interesting avenue would be to instead use mathematical models of supply chain dynamics. They are better designed for implementing global analytical sensitivity analyses, like those that have been already applied, for example, in population dynamics (Caswell 2001). The other interest is that the forest growth models could be incorporated into the same system of equations, given that these models are frequently based on the same mathematical formalism (like the one we used) (Wernsdörfer et al. 2012; Colin et al. 2017). By doing this, it would be possible to further perform novel global sensitivity analyses at the intersection between the ecological dimension and the economic dimension. For instance it would be possible to analyse the sensitivity of specific resource and supplies of bio-molecules in response to the perturbation of the demand for other bio-molecules or wood products.
Finally, regardless of how well we perform such sensitivity analyses, actors still need to evaluate whether some solutions would be more optimal or viable than others when considering other criteria. In this regard, several questions arise that would benefit from applying better optimisation algorithms. For instance, is it preferable that each industry strategically invest in its own specialised biorefinery? Or should they collectively invest in a big generic one that is strategically placed for everyone to contract with? Similar questions also arise for bio-energy conversion facilities (Elia and Floudas 2014; Thiriet et al. 2020). Then, once the biorefinery(ies) is(are) constructed, where should actors tactically establish the stocking sites to sustain the level of extractable bio-molecules inside the logs (Melo et al. 2009)? For such mid-term problems, viability theory algorithms are well suited to help minimise such risks under well defined constraints (Aubin 2009; Chavas 2015). Third, once the stocking sites are established, industrial actors need to sign contracts to supply by-products to one of the potential biorefineries. The nature of the contracts depends on many supply chain and ecological constraints. We think that Elia and Floudas (2014) and Rahmani et al. (2016) and Cambero and Sowlati (2016) present interesting optimisation tools for such complex multi-dimensional problems.
5 Conclusion
Before making decisions, supply chain actors with an interest in bio-molecules require specific background knowledge. It is not easy to obtain since it requires an extensive effort of scientific coordination to produce knowledge that spans different scales of observation, i.e. at the scales of bio-molecules, biomass components of trees, regional resources and, finally, the entire supply chain covering different transformation industries with their primary products and by-products, and other stakeholders. A major issue still is that adding new scales added more dimensions to the problem, making it harder to connect data, models and software and, ultimately, make sound decisions. Nevertheless, we demonstrated that once this connection was achieved, it was possible to draw a simple knowledge and decision pathway, containing a reduced set of steps. We demonstrated that the information produced for one step (scale) could be reasonably used to produce new information and solve a decision problem at another scale. We suggested efficient analytical, modelling and decision theory methods for those who want to optimise the decision process according to a diversity of socio-economic criteria. We hope that this study will help to design a more generic knowledge infrastructure and methodological framework, and that they could be used to solve a wide diversity of decision problems within the context of various species, resources and supply chains.
References
ADEME (2019) Schéma régional biomasse (srb) – rapport de diagnostic – région grand-est. https://www.grand-est.developpement-durable.gouv.fr207-lancement_acteurs_srb_gd_est.pdf
AGRESTE (2017) Chiffres et analyses en ligne pour la forêts, bois, industries du bois schéma. https://stats.agriculture.gouv.fr/disar-web/disaron/!searchurl/searchUiid/search.disar
Ananda J, Herath G (2009) A critical review of multi-criteria decision making methods with special reference to forest management and planning. Ecol Econ 68(10):2535–2548
Anttila A-K, Pirttilä A. M., Häggman H, Harju A, Venäläinen M, Haapala A, Holmbom B, Julkunen-Tiitto R (2013) Condensed conifer tannins as antifungal agents in liquid culture. Holzforschung 67(7):825–832
Aubin J-P (2009) Viability theory. Springer Science & Business Media
Bauer R, Billard A, Longuetaud F, Mothe F, Colin F (2019) Modelling the volume of bark from thickness measurement obtained by swedish gauge-application to the quantification of extractives. In: 21st International Nondestructive Testing and Evaluation of Wood Symposium
Baycheva-Merger T, Sotirov M (2020) The politics of an eu forest information system: Unpacking distributive conflicts associated with the use of forest information. Forest Policy and Economics 112:102110
Benković ET, Grohar T, žigon D, Švajger U, Janeš D, Kreft, S, Štrukelj B (2014) Chemical composition of the silver fir (abies alba) bark extract abigenol®;, and its antioxidant activity. Ind Crops Prod 52:23–28
Bergström D, Matisons M (2014) Forest refine, 2012-2014: efficient forest biomass supply chain management for biorefineries: synthesis report (no. 2014:18). Technical report
Bhardwaj K, Silva AS, Atanassova M, Sharma R, Nepovimova E, Musilek K, Sharma R, Alghuthaymi MA, Dhanjal DS, Nicoletti M et al (2021) Conifers phytochemicals: a valuable forest with therapeutic potential. Molecules 26(10):3005
Bianchi S, Koch G, Janzon R, Mayer I, Saake B, Pichelin F (2016) Hot water extraction of Norway spruce (picea abies [karst.]) bark: analyses of the influence of bark aging and process parameters on the extract composition. Holzforschung 70(7):619–631
Billard A, Bauer R, Mothe F, Jonard M, Colin F, Longuetaud F (2020) Improving aboveground biomass estimates by taking into account density variations between tree components. Ann For Sci 77 (4):1–18
Brennan M, Fritsch C, Cosgun S, Dumarcay S, Colin F, Gérardin P (2020a) Quantitative and qualitative composition of bark polyphenols changes longitudinally with bark maturity in Abies alba Mill. Ann For Sci 77(1):1–14
Brennan M, Fritsch C, Cosgun S, Dumarcay S, Colin F, Gérardin P (2020b) Yield and compositions of bark phenolic extractives from three commercially significant softwoods show intra-and inter-specific variation. Plant Physiol Biochem 155:346–356
Brennan M, Hentges D, Cosgun S, Dumarcay S, Colin F, Gérardin C, Gérardin P (2020c) Intraspecific variability of quantity and chemical composition of ethanolic knotwood extracts along the stems of three industrially important softwood species: Abies alba, picea abies and pseudotsuga menziesii. Holzforschung 1(ahead-of-print)
Brunet-Navarro P, Jochheim H, Muys B (2016) Modelling carbon stocks and fluxes in the wood product sector: a comparative review. Glob Change Biol 22(7):2555–2569
Cambero C, Sowlati T (2016) Incorporating social benefits in multi-objective optimization of forest-based bioenergy and biofuel supply chains. Appl Energy 178:721–735
Carter CR, Rogers DS, Choi TY (2015) Toward the theory of the supply chain. J Supply Chain Manag 51(2):89–97
Caswell H (2001) Matrix population models construction, analysis, and interpretation. Sunderland Massachusetts Sinauer Associates
Cencic O (2016) Nonlinear data reconciliation in material flow analysis with software stan. Sustain Environ Res 26:291–298
Chavas J-P (2015) Dynamics, viability, and resilience in bioeconomics. Annu Rev Resour Econ 7(1):209–231
Colin A, Wernsdörfer H, Thivolle-Cazat A, Jean-Daniel B (2017) Forest inventory-based projection Systems for Wood and Biomass Availability, vol. 29, chapter 13. Springer
D’Amato D, Droste N, Allen B, Kettunen M, Lähtinen K, Korhonen J, Leskinen P, Matthies BD, Toppinen A (2017) Green, circular, bio economy: a comparative analysis of sustainability avenues. J Clean Prod 168:716–734
Dedrie M, Jacquet N, Bombeck P-L, Hébert J, Richel A (2015) Oak barks as raw materials for the extraction of polyphenols for the chemical and pharmaceutical sectors: a regional case study. Ind Crop Prod 70:316–321
Elia JA, Floudas CA (2014) Energy supply chain optimization of hybrid feedstock processes: a review. Ann Rev Chem Biomol Eng 5:147–179
Ficko A, Poljanec A, Boncina A (2011) Do changes in spatial distribution, structure and abundance of silver fir (Abies alba Mill.) indicate its decline? For Ecol Manag 261(4):844–854
Genet A, Wernsdörfer H, Jonard M, Pretzsch H, Rauch M, Ponette Q, Nys C, Legout A, Ranger J, Vallet P, Saint-André L (2011) Ontogeny partly explains the apparent heterogeneity of published biomass equations for fagus sylvatica in central europe. For Ecol Manag 261:1188–1202
Gómez-García E, Biging G, García-Villabrille JD, Crecente-Campo F, Castedo-Dorado F, Rojo-Alboreca A (2015) Cumulative continuous predictions for bole and aboveground woody biomass in eucalyptus globulus plantations in northwestern spain. Biomass Bioenergy 77:155–164
Gong G, Guan Y-Y, Zhang Z-L, Rahman K, Wang S-J, Zhou S, Luan X, Zhang H (2020) Isorhamnetin: a review of pharmacological effects. Biomed Pharma 128:110301
Hassegawa M, Gélinas N, Beaudoin D, Achim A (2018) Assessing the potential impact of a biorefinery product from sawmill residues on the profitability of a hardwood value chain. Can J For Res 48(8):857–868
Holmbom B (2011) Extraction and utilisation of non-structural wood and bark components. Biorefin Forest Resour:178–224
Holmbom B, Eckerman C, Eklund P, Hemming J, Nisula L, Reunanen M, Sjöholm R, Sundberg A, Sundberg K, Willför S (2003) Knots in trees–a new rich source of lignans. Phytochem Rev 2(3):331–340
Hounzandji API, Jonard M, Nys C, Saint-André L, Ponette Q (2014) Improving the robustness of biomass functions: from empirical to functional approaches. Ann For Sci 72:795–810
IGN (2018) Mémento de l’inventaire forestier. http://www.ign.fr/institut/kiosque/memento-linventaire-forestier
Jablonsky M, Nosalova J, Sladkova A, Haz A, Kreps F, Valka J, Miertus S, Frecer V, Ondrejovic M, Sima J et al (2017) Valorisation of softwood bark through extraction of utilizable chemicals. a review. Biotechnol Adv 35(6):726–750
Jayasinghe P, Hawboldt K (2012) A review of bio-oils from waste biomass: Focus on fish processing waste. Renew Sustain Energy Rev 16(1):798–821
Jyske T, Laakso T, Latva-Mäenpää H, Tapanila T, Saranpää P (2014) Yield of stilbene glucosides from the bark of young and old Norway spruce stems. Biomass Bioenergy 71:216–227
Kangas A, Räty M, Korhonen KT, Vauhkonen J, Packalen T (2019) Catering information needs from global to local scales—potential and challenges with national forest inventories. Forests 10(9):800
Kebbi-Benkeder Z, Colin F, Dumarċay S, Gérardin P (2015) Quantification and characterization of knotwood extractives of 12 european softwood and hardwood species. Ann Forest Sci 72(2):277–284
Kebbi-Benkeder Z, Manso R, Gérardin P, Dumarċay S, Chopard B, Colin F (2017) Knot extractives: a model for analysing the eco-physiological factors that control the within and between-tree variability. Trees 31(5):1619–1633
Kilulya KF, Msagati TA, Mamba BB, Ngila JC, Bush T (2014) Effect of site, species and tree size on the quantitative variation of lipophilic extractives in eucalyptus woods used for pulping in south africa. Ind Crop Prod 56:166–174
Köhl M, Magnussen SS, Marchetti M (2006) Sampling methods, remote sensing and GIS multiresource forest inventory. Springer Science & Business Media
Kopaczyk JM, Warguła J, Jelonek T (2020) The variability of terpenes in conifers under developmental and environmental stimuli. Environ Exper Botany:104197
Lacoste C, Čop M, Kemppainen K, Giovando S, Pizzi A, Laborie M-P, Sernek M, Celzard A (2015) Biobased foams from condensed tannin extracts from Norway spruce (picea abies) bark. Ind Crop Prod 73:144–153
Lenglet J, Courtonne J-Y, Caurla S (2017) Material flow analysis of the forest-wood supply chain: a consequential approach for log export policies in france. J Clean Prod 165:1296–1305
Lewinsohn E, Gijzen M, Croteau R (1991) Defense mechanisms ovf conifers: differences in constitutive and wound-induced monoterpene biosynthesis among species. Plant Physiol 96(1):44–49
Liao Y, Koelewijn S-F, Van den Bossche G, Van Aelst J, Van den Bosch S, Renders T, Navare K, Nicolaï T, Van Aelst K, Maesen M et al (2020) A sustainable wood biorefinery for low–carbon footprint chemicals production. Science 367(6484):1385–1390
Lindner M, Werhahn-Mees W, Suominen T, Vötter D, Zudin S, Pekkanen M, Päivinen R, Roubalova M, Kneblik P, Brüchert F et al (2012) Conducting sustainability impact assessments of forestry-wood chains: examples of tosia applications. Eur J For Res 131(1):21–34
Longuetaud F, Mothe F, Fournier M, Dlouha J, Santenoise P, Deleuze C (2016) Within-stem maps of wood density and water content for characterization of species: a case study on three hardwood and two softwood species. Ann Forest Sci 73(3):601–614
Mäkelä A, Valentine H (2020) Models of tree and stand dynamics: Theory, formulation and application. Models of Tree and Stand Dynamics
Martin BT (2009) Opportunities for an Online GIS-based Wood Supply Management System. PhD thesis, Virginia Tech
Melo MT, Nickel S, Saldanha-Da-Gama F (2009) Facility location and supply chain management–a review. Eur J Oper Res 196(2):401–412
Min H, Zhou G (2002) Supply chain modeling: past, present and future. Comput Indust Eng 43(1-2):231–249
Mukhopadhyay AK (2006) Antioxidants-natural and Synthetic. Amani Int’l Publishers
Nabarlatz D, Ebringerová A, Montané D (2007) Autohydrolysis of agricultural by-products for the production of xylo-oligosaccharides. Carbohyd Polym 69(1):20–28
Naik SN, Goud VV, Rout PK, Dalai AK (2010) Production of first and second generation biofuels: a comprehensive review. Renew Sustain Energy Rev 14(2):578–597
Näyhä A, Hämäläinen S, Pesonen H (2011) Forest biorefineries–a serious global business opportunity. Biorefining of forest resources. University of jyväskylä, Laboratory of Applied Chemistry. Paper Engineers’ Association, Helsinki, Finland, pp 132–150
Nisula L (2018) Wood extractives in conifers: a study of stemwood and knots of industrially important species. Åbo Akademis förlag-Åbo Akademi University Press
Normandin D (1990) Le chêne français et ses produits dérivés. Marché intérieur et concurrence internationale. Revue forestière française XLII, pp 110–118
Paquet P, Deroubaix G (2003) Extension de l’éligibilité de la séquestration forestière du carbone à l’ensemble des stocks de la filière bois. CTBA Coordination, ADEME, GICC Programme
Philp J (2018) The bioeconomy, the challenge of the century for policy makers. Biotechnol 40:11–19
Pichancourt JB (2020) Data and code to estimate the quantity of molecular extractives along the forestry supply-chain for the species Abies alba in the Grand-Est region, France. [dataset], Data INRAE repository,V2. https://doi.org/10.15454/3KESWN
Pichancourt J. -B., Manso R, Ningre F, Fortin M (2018) A carbon accounting tool for complex and uncertain greenhouse gas emission life cycles. Environ Modell Softw 107:158–174
Pietarinen S, Willför SM, Sjöholm RE, Holmbom B (2005) Bioactive phenolic substances in important tree species. part 3: Knots and stemwood of acacia crassicarpa and a. mangium. Holzforschung 59 (1):94–101
Piispanen R, Willför S, Saranpää P, Holmbom B (2008) Variation of lignans in Norway spruce (picea abies [l.] karst.) knotwood: within-stem variation and the effect of fertilisation at two experimental sites in finland. Trees 22(3):317–328
Pizzi A (2019) Tannins: Prospectives and actual industrial applications. Biomolecules 9(8):344
Rabelo L, Sarmiento AT, Jones A (2011) Stability of the supply chain using system dynamics simulation and the accumulated deviations from equilibrium. Modelling and Simulation in Engineering 2011
Rahmani Y, Ramdane Cherif-Khettaf W, Oulamara A (2016) The two-echelon multi-products location-routing problem with pickup and delivery: formulation and heuristic approaches. Int J Prod Res 54(4):999–1019
Rivoire M, Longuetaud F, Deleuze C, Saint André L, Vallet P, Morneau F, Bouvet A, Gauthier A (2010) Exploring the variability of biomass distribution in individual forest trees. In: XXIII IUFRO World Congress. Forests for the Future: Sustaining Society and the Environment, pp 1
Roitto M, Siwale W, Tanner J, Ilvesniemi H, Julkunen-Tiitto R, Verkasalo E (2016) Characterization of extractives in tree biomass and by-products of plywood and saw mills from finnish birch in different climatic regions for value-added chemical products. In: Wood Science-Annual Meeting, pp 190
Routa J, Brännström H, Anttila P, Mäkinen M, Jänis J, Asikainen A (2017) Wood extractives of finnish pine, spruce and birch – availability and optimal sources of compounds
Royer M, Houde R, Stevanovic T (2010) Potentiel de développement lié aux extractibles forestiers: État des connaissances et revue des marchés. volet 1: Les extractibles forestiers québecois. Technical report
Royer M, Houde R, Stevanovic T (2013) Non-wood forest products based on extractives-a new opportunity for canadian forest industry part 2-softwood forest species. J Food Res 2(5):164
Royer M, Houde R, Viano Y, Stevanovic T (2012) Non-wood forest products based on extractives-a new opportunity for the canadian forest industry part 1: Hardwood forest species. J Food Res 1(3):8
Rustad T, Storrø I, Slizyte R (2011) Possibilities for the utilisation of marine by-products. Int J Food Sci Technol 46(10):2001–2014
Santana MAE, Okino EYA (2007) Chemical composition of 36 brazilian amazon forest wood species. Holzforschung 61(5):469–477
Sekine N, Shibutani S, Yatagai M (2013) Chemical composition of the terpenoids in wood and knots of abies species. Eur J Wood Wood Prod 71(5):679–682
Sinclair G, Dymond D (1973) The distribution and composition of extractives in jack pine trees. Can J For Res 3(4):516–521
Stevanovic T, Diouf PN, Garcia-Perez ME (2009) Bioactive polyphenols from healthy diets and forest biomass. Curr Nutrition Food Sci 5(4):264–295
Thiriet P, Bioteau T, Tremier A (2020) Optimization method to construct micro-anaerobic digesters networks for decentralized biowaste treatment in urban and peri-urban areas. J Clean Prod 243:118478
Vahid S, Lehoux N, de Santa-Eulalia LA, D’Amours S, Frayret J-M, Venkatadri U (2016) Supply chain modelling frameworks for forest products industry: a systematic literature review. INFOR: Inf Syst Oper Res 54(1):52–75
Valette N, Perrot T, Sormani R, Gelhaye E, Morel-Rouhier M (2017) Antifungal activities of wood extractives. Fungal Biol Rev 31(3):113–123
Velema G (2004) Management and benefits of pulp and paper mill residuals at domtar cornwall. Pulp Paper Can 105(7):26–32
Verkasalo E, Möttönen V, Roitto M, Vepsäläinen J, Kumar A, Ilvesniemi H, Siwale W, Julkunen-Tiitto R, Raatikainen O, Sikanen L (2021) Extractives of stemwood and sawmill residues of scots pine (pinus sylvestris l.) for biorefining in four climatic regions in finland—phenolic and resin acid compounds. Forests 12(2):192
Vidal C, Alberdi I, Hernández L., Redmond J (2016) National forest inventories: Assessment of Wood Availability and Use. Springer
Vieira AA, Dias LM, Santos MY, Pereira GA, Oliveira JA (2020) Supply chain data integration: a literature review. Journal of Industrial Information Integration, pp 100161
Weiskittel AR, Hann DW, Kershaw Jr JA, Vanclay JK (2011) Forest growth and yield modeling. Wiley
Wernsdörfer H, Colin A, Bontemps J-D, Chevalier H, Pignard G, Caurla S, Leban J-M, Hervé J-C, Fournier M (2012) Large-scale dynamics of a heterogeneous forest resource are driven jointly by geographically varying growth conditions, tree species composition and stand structure. Ann For Sci 69(7):829–844
Willför S, Hemming J, Reunanen M, Eckerman C, Holmbom B (2003) Lignans and lipophilic extractives in Norway spruce knots and stemwood. Holzforschung 57(1):27–36
Willför S, Nisula L, Hemming J, Reunanen M, Holmbom B (2004a) Bioactive phenolic substances in industrially important tree species. part 1: Knots and stemwood of different spruce species. Holzforschung 58(4):335–344
Willför S, Nisula L, Hemming J, Reunanen M, Holmbom B (2004b) Bioactive phenolic substances in industrially important tree species. part 2: Knots and stemwood of fir species. Holzforschung 58 (6):650–659
Willför S, Reunanen M, Eklund P, Sjöholm R, Kronberg L, Fardim P, Pietarinen S, Holmbom B (2004c) Oligolignans in Norway spruce and scots pine knots and Norway spruce stemwood. Holzforschung 58(4):345–354
Willför S, Sundberg A, Rehn P, Holmbom B, Saranpää P (2005) Distribution of lignans in knots and adjacent stemwood of picea abies. Holz Roh-und Werkstoff 63(5):353–357
Acknowledgements
We are grateful to Frédéric Mothe, Gail Wagman and three anonymous reviewers for discussions, comments and suggestions of corrections on earlier versions of this manuscript.
Funding
ExtraFor-Est is a 1.55 million euros research projet that was managed by UMR 1434 Silva and EA 4370 LERMAB. It has been supported by the following funding agencies: the French Ministry of Agriculture (contractual project EXTRAFOR-EST/MAAF), the Lorraine-FEDER (contractual project EXTRAFOR-EST/FEDER), the French National Research Agency (ANR) as part of the Investissements d’Avenir programme (ANR-11-LABX-0002-01), the Lab of Excellence ARBRE (contractual project BARK-TAN-BIO). ADEME and Grand Est Region supported a PhD thesis on basic density of wood tissues.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All interviews of human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent for publication
The authors gave their consent for publication.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Erwin Dreyer
Author contribution
Francis Colin is the lead PI of the EXTRAFOREST project. Philippe Gerardin, Stéphane Dumarçay, Maree Brennan and Sylvain Coşgun designed the chemical experiment and produced the molecular results for all tree tissues, and David Jean Hentges performed the analysis. Fleur Longuetaud, Antoine Billard & Rodolphe Bauer designed the experiment and analysed the dataset and produced dendrometric results for all tree tissues and at the scale of the tree. Antoine Colin and Henri Cuny designed the GIS model and Holger Wernsdörfer the forest growth model, and Henri Cuny produced the map of the diversity and quantity of extractives. Antoine Colin, Henri Cuny, Jean-Baptiste Pichancourt and Mathieu Fortin worked on the connection between the forest growth model and the supply chain model. Loïc Karwadec, Sylvain Caurla and Holger Wernsdörfer designed the socio-economic interviews and Loïc Kerwadec interviewed forestry actors to get relevant quantitative and qualitative data to improve the supply chain model. Jean-Baptiste Pichancourt designed the supply chain model for the Grand Est region and produced the results. Jean-Baptiste Pichancourt designed the scientific study and is the main writer. All co-authors contributed to the writing.
Code and data availability
Original data and code of models of the current study are available at the Data INRAE repository, https://doi.org/10.15454/3KESWN
Consent to participate
The authors gave their consent to participate in this study.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pichancourt, JB., Bauer, R., Billard, A. et al. A generic information framework for decision-making in a forest-based bio-economy. Annals of Forest Science 78, 97 (2021). https://doi.org/10.1007/s13595-021-01110-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-021-01110-y