Abstract
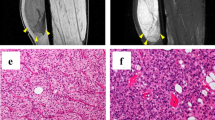
Pleomorphic liposarcoma (PLPS) is a highly malignant subtype of liposarcoma. It is histologically characterized by the presence of pleomorphic lipoblasts and can be accompanied by morphological foci that demonstrate differentiation to other histological lineages. PLPS is rare and accounts for only 5% of all liposarcomas. PLPS exhibits poor prognosis; distant metastases develop in 30–50% of patients after curative surgical resection, tumor-associated mortality occurs in up to 50% of patients, and effective chemotherapies for PLPS have not been established. The histological accompaniment of other morphological foci is an important prognostic factor for PLPS, and the development of chemotherapies for PLPS considering the histological morphology is necessary. Patient-derived cancer cell lines are critical tools for basic and pre-clinical research to understand diseases and develop chemotherapies. However, only two PLPS-derived cell lines have been reported, and their donor tumor specimens did not histologically accompany morphological foci other than lipoblasts. Thus, there is a need to establish patient-derived PLPS cell lines from various histological morphologies. Here, we report a novel PLPS cell line from a tumor specimen that histologically accompanied pleomorphic and bone-forming foci, and named it NCC-PLPS2-C1. NCC-PLPS2-C1 cells demonstrated constant proliferation, spheroid formation, and invasion capability in vitro. Screening of antitumor agents in NCC-PLPS2-C1 cells showed that bortezomib, romidepsin, and trabectedin were effective against NCC-PLPS2-C1. In conclusion, we report the first PLPS cell line from a tumor specimen that was morphologically accompanied by pleomorphic and born-forming foci. We believe that NCC-PLPS2-C1 will be useful for the development of novel chemotherapies for PLPS.




Similar content being viewed by others
References
Gjorgova-Gjeorgjievski S, Thway K, Dermawan JK, et al. Pleomorphic liposarcoma: a series of 120 cases with emphasis on morphologic variants. Am J Surg Pathol. 2022;46:1700–5.
Gebhard S, Coindre JM, Michels JJ, et al. Pleomorphic liposarcoma: clinicopathologic, immunohistochemical, and follow-up analysis of 63 cases: a study from the French Federation of Cancer Centers Sarcoma Group. Am J Surg Pathol. 2002;26:601–16.
Lee ATJ, Thway K, Huang PH, Jones RL. Clinical and molecular spectrum of liposarcoma. J Clin Oncol. 2018;36:151–9.
Wang L, Luo R, Xiong Z, Xu J, Fang D. Pleomorphic liposarcoma: an analysis of 6 case reports and literature review. Med (Baltim). 2018;97: e9986.
Fritz B, Schubert F, Wrobel G, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–8.
Idbaih A, Coindre JM, Derré J, et al. Myxoid malignant fibrous histiocytoma and pleomorphic liposarcoma share very similar genomic imbalances. Lab Invest. 2005;85:176–81.
Rieker RJ, Joos S, Bartsch C, et al. Distinct chromosomal imbalances in pleomorphic and in high-grade dedifferentiated liposarcomas. Int J Cancer. 2002;99:68–73.
Adachi T, Oda Y, Sakamoto A, et al. Prognostic factors in the so-called malignant mesenchymoma: a clinicopathological and immunohistochemical analysis. Oncol Rep. 2003;10:803–11.
Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–50.
Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–6.
Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–84.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Warren A, Chen Y, Jones A, et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat Commun. 2021;12:22.
Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38.
Hideyuki T, Yoko T, Daisuke M, Miharu K, Tohru M, Hiroshi M. Cell line individualization by str multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tissue Cult Res Commun. 1999;18:329–38.
Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci USA. 2001;98:8012–7.
Drexler HG, Dirks WG, MacLeod RA, Uphoff CC. False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017;140:1209–14.
Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7.
Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: a hospital-based study. Cancer Sci. 2019;110:1480–90.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Cox C, Teknos TN, Barrios M, Brewer GJ, Dick RD, Merajver SD. The role of copper suppression as an antiangiogenic strategy in head and neck squamous cell carcinoma. Laryngoscope. 2001;111:696–701.
Schöffski P. Established and experimental systemic treatment options for advanced liposarcoma. Oncol Res Treat. 2022;45:525–43.
Wabitsch M, Brüderlein S, Melzner I, Braun M, Mechtersheimer G, Möller P. LiSa-2, a novel human liposarcoma cell line with a high capacity for terminal adipose differentiation. Int J Cancer. 2000;88:889–94.
Noguchi R, Yoshimatsu Y, Ono T, et al. Establishment and characterization of NCC-PLPS1-C1, a novel patient-derived cell line of pleomorphic liposarcoma. Hum Cell. 2021;34:688–97.
Gao P, Seebacher NA, Hornicek F, Guo Z, Duan Z. Advances in sarcoma gene mutations and therapeutic targets. Cancer Treat Rev. 2018;62:98–109.
Czarnecka AM, Synoradzki K, Firlej W, et al. Molecular biology of osteosarcoma. Cancers (Basel). 2020;12:2130.
Liu J, Zhao R, Jiang X, Li Z, Zhang B. Progress on the application of bortezomib and bortezomib-based nanoformulations. Biomolecules. 2021;12:51.
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:1414.
Zarin DA, Fain KM, Dobbins HD, Tse T, Williams RJ. 10-Year update on study results submitted to ClinicalTrials.gov. N Engl J Med. 2019;381:1966–74.
Demetri GD, von Mehren M, Jones RL, et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: results of a phase III randomized multicenter clinical trial. J Clin Oncol. 2016;34:786–93.
Acknowledgements
We thank Drs. E. Kobayashi, K. Ogura, S. Osaki, S. Fukushima, K. Sato, S. Ishihara, Y. Toda (Department of Musculoskeletal Oncology, the National Cancer Center Hospital) for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical support provided by Mrs. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for providing English language editing services and for their constructive comments on this manuscript. This study was technically assisted by the Fundamental Innovative Oncology Core of the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (grant number: 20ck0106537h0003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethics approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004-050). Animal experiments were conducted in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute.
Informed consent
Written informed consent was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2022_828_MOESM1_ESM.tiff
Supplementary Fig. 1 Short tandem repeat patterns of NCC-PLPS2-C1 cells and original tumor tissue. (A) Short tandem repeat patterns of NCC-PLPS2-C1 cells (p20) and (B) short tandem repeat patterns of original tumor tissue of NCC-PLPS2-C1 (TIFF 3922 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akiyama, T., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-PLPS2-C1: a novel cell line of pleomorphic liposarcoma. Human Cell 36, 468–475 (2023). https://doi.org/10.1007/s13577-022-00828-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00828-9