Abstract
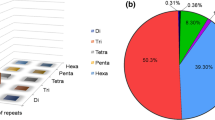
Okra (Abelmoschus esculentus) is an important nutritious vegetable. Despite its high economic and industrial value, very little attention has been paid to assess genetic diversity of okra at molecular level. For effective conservation and proper deployment of germplasm, a study on diversity analysis of okra germplasm was conducted with DNA markers. Microsatellite/Simple sequence repeat (SSR) markers were utilized to evaluate the genetic diversity among 96 accessions of Abelmoschus, of which 92 accessions were of A. esculentus and one accession each of A. tuberculatus, A. moschatus, A. moschatus subspecies tuberosus and A. manihot. A set of 40 SSR primers were tested, of which 30 primers gave reproducible amplification which were used further for diversity analysis. With a mean of 7.1 bands per SSR, DNA amplification with 30 SSRs generated a total 213 bands, of which 60.66 % were recorded polymorphic. Polymorphic information content ranged between 0.11 and 0.80 with an average of 0.52, indicating that the majority of primers were informative. The Jaccard’s coefficient ranged from 0.107 to 0.969. The UPGMA analysis grouped Abelmoschus genotypes into three main clusters at a cut-off of 0.20. Results of present study revealed that sufficient variation exists among the studied accessions and GAO-5 which was found highly diverse can be exploited for okra improvement. The outcome of present research would assist to make use of Ablemoschus germplasm for okra breeding.


Similar content being viewed by others
Abbreviations
- EST:
-
Expressed sequence tag
- SSR:
-
Simple sequence repeat
- RAPD:
-
Random amplified polymorphic DNA
- ISSR:
-
Inter simple sequence repeat
- SRAP:
-
Sequence-related amplified polymorphism
References
Adelakun OE, Oyelade OJ, Ade-Omowaye BIO, Adeyemi IA, Van de Venter M, Koekemoer TC (2009) Influence of pretreatment on yield, chemical and antioxidant properties of a Nigerian okra seed (Abelmoschus esculentus Moench.) flour. Food Chem Toxicol 47:657–661
Akash MW, Shiyab SM, Saleh MI (2013) Yield and AFLP analyses of inter-landrace variability in okra (Abelmoschus esculentus L.). Life Sci J 10:2771–2779
Aladele SE (2009) Morphological distinctiveness and metroglyph analysis of fifty accessions of West African okra (Abelmoschus caillei) (A. Chev.) Stevels. J Plant Breed Crop Sci 1:273–280
Aladele SE, Ariyo OJ, de La Pena R (2008) Genetic relationships among West African okra (Abelmoschus caillei) and Asian genotypes (Abelmoschus esculentus) using RAPD. Afr J Biotechnol 7:1426–1431
Ariyo OJ (1993) Genetic diversity in West African Okra [Abelmoschus caillei (A. Chev.) Stevels] multivariate analysis of morphological and agronomic characteristics. Genet Resour Crop Evol 40:25–32
Bisht IS, Bhat KV (2006) Okra (Abelmoschus spp.). In: Singh RJ (ed) Genetic resources, chromosome engineering and crop improvement: vegetable crops, vol 3. CRC Press, Boca Raton, pp 148–162
Calisir S, Yildiz MU (2005) A study on some physico-chemical properties of Turkeyokra (Hibiscus esculenta) seeds. J Food Eng 68:73–78
Datta PC, Naug A (1968) A few strains of Abelmoschus esculentus (L.) Moench. Their karyological study in relation to phylogeny and organ development. Beitr Biol Pflanzen 45:113–126
Fougat RS, Purohit AR, Kumar S, Parekh MJ, Kumar M (2015) Study of SSR based genetic diversity in Abelmoschus species. Indian J Agric Sci 85(9):1223–1228
Giese J (2014) Industrial applications of selected JFS articles. J Food Sci 79:4–5
Gulsen O, Karagul S, Abak K (2007) Diversity and relationships among Turkish okra germplasm by SRAP and phenotypic marker polymorphism. Biol Bratisl 62:41–45
Hamon S, Koechlin J (1991) The reproductive biology of okra. 2. Self-fertilization kinetics in the cultivated okra (Abelmoschus esculentus), and consequences for breeding. Euphytica 53:49–55
Hamon S, van Sloten DH (1995) Okra. In: Smartt J, Simmonds NW (eds) Evolution of crop plants. Wiley, New York, pp 350–357
Haq I, Khan AA, Azmat MA (2013) Assessment of genetic diversity in okra (Abelmoschus esculentus L.) using RAPD markers. Pak J Agric Sci 50:655–662
Kaur K, Pathak M, Kaur S, Pathak D, Chawla N (2013) Assessment of morphological and molecular diversity among okra (Abelmoschus esculentus (L.) Moench.) germplasm. Afr J Biotechnol 12:3160–3170
Kochko AD, Hamon S (1990) A rapid and efficient method for the isolation of restrictable total DNA from plants of the genus Abelmoschus. Plant Mol Biol Rep 8:3–7
Kumar S, Dagnoko S, Haougui A, Ratnadass A, Pasternak D, Kouame C (2010) Okra (Abelmoschus spp.) in west and central Africa: potential and progress on its improvement. Afr J Agric Res 5:3590–3598
Kyriakopoulou OG, Arens P, Pelgrom KTB, Bebeli P, Passam HC (2014) Genetic and morphological diversity of okra (Abelmoschus esculentus [L.] Moench.) genotypes and their possible relationships, with particular reference to Greek landraces. Sci Hortic 171:58–70
Martinello GE, Leal NR, Amaral AT Jr, Pereira MG, Daher RF (2001) Comparison of morphological characteristics and RAPD for estimating genetic diversity in Abelmoschus spp. Acta Hortic 546:101–104
Mortz E, Krogh TN, Vorum H, Gorg A (2001) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359–1363
Noorlaila A, Siti A, Asmeda R, Norizzah AR (2015) Emulsifying properties of extracted okra (Abelmoschus esculentus L.) mucilage of different maturity index and its application in coconut milk emulsion. Int Food Res J 22:782–787
Noormohammadi AZ, Taghavi AE, Foroutan BM, Sheidai BM, Alishah CO (2013) Structure analysis of genetic diversity in tetraploid and diploid cotton genotypes. Int J Plant Anim Environ Sci 3:79–90
NRCNAs (2006) “Okra”. Lost Crops of Africa: vol II: Vegetables. Lost Crops of Africa. 2. National Academies Press. ISBN 978-0-309-10333-6
Oliveira KM, Pinto LR, Marconi TG, Mollinari M, Ulian EC, Chabregas SM, Falco MC, Burnquist W, Garcia AAF, Souza AP (2009) Characterization of new polymorphic functional markers for sugarcane. Genome 52:191–209
Omonhinmin CA, Osawaru ME (2005) Morphological characterisation of two species of Abelmoschus: Abelmoschus esculentus and Abelmoschus caillei. Plant Genet Res 144:51–55
Oppong-Sekyere D (2011) Assessment of genetic diversity in a collectionof Ghanaian okra germplasm (Abelmoschus spp. L.) using morphological markers. MSc. thesis presented to the Department of Crop Science, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana, pp 42–60
Osawaru ME, Ogwu MC, Dania-Ogbe FM (2013) Morphological assessment of the genetic variability among 53 accessions of West African okra [Abelmoschus caillei (A. Chev.) Stevels] from South Western Nigeria. Niger J Basic Appl Sci 21:227–238
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Poehlman JM, Sleper DA (1995) Breeding field crops, 4th edn. Iowa State University Press, Ames
Prakash K, Pitchaimutu M, Ravishankar KV (2011) Assessment of genetic relatedness among okra genotypes [Abelmoschus esculentus (L.) Moench.] using RAPD markers. Electron J Plant Breed 2:80–86
Ramneek Jaskirat, Pathak M, Pathak D (2012) Preliminary investigation on the cross transferability of cotton SSR markers to Abelmoschus species. Crop Improv 39:80–84
Rohlf FJ (1998) NTSYSpc: numerical taxonomy and multivariate analysis system. Version 2.02. Exeter Software, Setauket
Saifullah M, Rabbani MG, Garvey EJ (2010) Estimation of genetic diversity of okra (Abelmoschus esculentus L. Moench) using RAPD markers. SAARC J Agric 8:19–28
Sawadogo M, Ouedraogo JT, Balma D, Ouedraogo M, Gowda BS, Botanga C, Timko MP (2009) The use of cross species SSR primers to study genetic diversity of okra from Burkina Faso. Afr J Biotechnol 8:2476–2482
Schafleitner R, Kumar S, Lin C, Hegde SG, Ebert A (2013) The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 517:27–36
Sutar S, Patil P, Aitawade M, John J, Malik S, Rao SR, Yadav S, Bhat KV (2013) A new species of Abelmoschus Medik. (Malvaceae) from Chhattisgarh. India Genet Resour Crop Evol 60:1953–1958
Torkpo SK, Danquah EY, Offei SK, Blay ET (2006) Esterase, total protein and seed storage protein diversity in Okra (Abelmoschus esculentus L. Moench). West Afr J Appl Ecol 9:1–7
Werner O, Magdy M, Ros RM (2015) Molecular systematics of Abelmoschus (Malvaceae) and genetic diversity within the cultivated species of this genus based on nuclear ITS and chloroplast rpL16 sequence data. Genet Resour Crop Evol. doi:10.1007/s10722-015-0259-x
Yap IV, Nelson RJ (1996) Winboot: a program for performing bootstrap analysis of binary data to determine the confidence of UPGMA-based dendrograms. IRRI, Manilla
Yuan CY, Zhang C, Wang P, Hu S, Chang HP, Xiao WJ, Lu XT, Jiang SB, Ye JZ, Guo XH (2014) Genetic diversity analysis of okra (Abelmoschus esculentus L.) by inter-simple sequence repeat (ISSR) markers. Genet Mol Res 13:3165–3175
Author contribution
S.K. and M.J.P performed extraction of the DNA. C.B.P. and S.K.P. carried out polymerase chain reactions. M.K. and S.K. analysed the data. B.R.P. provided the seed material and manage the seedling growth. S.K. and R.S.F. contributed by designing the experiment, supervising the whole experiment and preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
SSR based dendrogram of the genetic similarities among twenty five accessions of Abelmoschus species achieved by the UPGMA method. The bootstrap values which are higher than 30 are shown. (PPTX 227 kb)
Rights and permissions
About this article
Cite this article
Kumar, S., Parekh, M.J., Fougat, R.S. et al. Assessment of genetic diversity among okra genotypes using SSR markers. J. Plant Biochem. Biotechnol. 26, 172–178 (2017). https://doi.org/10.1007/s13562-016-0378-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-016-0378-2