Abstract
Purpose
To clarify the ambiguity of the function of CMTM3 in the development of hepatocellular carcinoma (HCC) and explore its molecular mechanism.
Methods
The Cmtm3-KO C57BL/6 mouse strain was established using CRISPR-Cas9. Acute liver damage and HCC models were induced by peritoneal injection of 100 or 25 mg/kg.BW N-Nitrosodiethylamine (DEN) to male mice. Liver function and histology were evaluated by blood serum levels of AST and ALT, and HE staining. Gene and protein expression in liver tissues was investigated by RNA-seq, RT-qPCR, Western blotting, immunohistochemistry, and immunofluorescence. Protein–protein interactions were studied by STRING and topological measures. The mRNA expression of CMTM3 and PPARs and patient survival were analyzed using the UALCAN database.
Results
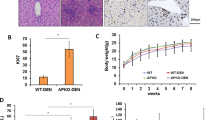
Global knockout of Cmtm3 in KO mice was successfully confirmed. Cmtm3 knockout alleviated DEN-induced acute damage to liver histological integrity and liver function, reduced DNA damage and apoptosis, and also caused a significantly reduced number (WT: 8.7 ± 5.5 vs. KO: 2.7 ± 3.1, P = 0.0394) and total size of tumors (WT: 130.9 ± 181.8 mm2 vs. KO: 9.3 ± 11.5 mm2, P = 0.026) in the liver. Mechanistically, Cmtm3 knockout resulted in reduced expression and inactivation of Pparγ and its downstream lipid metabolism genes (e.g. Adipoq) upon DEN intoxication. CMTM3 and PPARγ were both overexpressed in HCC, and higher levels of both genes were associated with worse overall survival of HCC patients.
Conclusion
This study clarified the pro-tumorigenesis role of CMTM3 in HCC in vivo, possibly through the upregulation of PPARγ and activation of the PPAR pathway.
Graphical abstract







Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. RNA-seq dataset is available at Sequence Read Archive (accession no. PRJNA850897).
Abbreviations
- BP:
-
Biological process
- CC:
-
Cellular component
- CKLFSF:
-
Chemokine-like factor superfamily
- CMTM3:
-
CKLF-like MARVEL transmembrane domain-containing member 3
- CRISPR-Cas9:
-
Clustered regularly interspaced short palindromic repeats-associated protein 9
- CYP2E1:
-
Cytochrome P450 2E1
- DEN:
-
N-nitrosodiethylamine
- GO:
-
Gene ontology
- HCC:
-
Hepatocellular carcinoma
- HE:
-
Hematoxylin-eosin
- IHC:
-
Immunohistochemistry
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KO:
-
Knock out
- KOG:
-
EuKaryotic Ortholog Groups
- MF:
-
Molecular function
- PCR:
-
Polymerase chain reaction
- PPAR:
-
Peroxisome proliferator-activated receptor
- PPI:
-
Protein-protein interaction
- RNA-seq:
-
Whole transcriptome sequencing
- SD:
-
Standard deviation
- TUNEL:
-
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling
- VEGF:
-
Vascular endothelial growth-factor
- WT:
-
Wild type
References
W. Han, P. Ding, M. Xu, L. Wang, M. Rui, S. Shi, Y. Liu, Y. Zheng, Y. Chen, T. Yang, D. Ma, Identification of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 81, 609–617 (2003)
J. Zhong, Y. Wang, X. Qiu, X. Mo, Y. Liu, T. Li, Q. Song, D. Ma, W. Han, Characterization and expression profile of CMTM3/CKLFSF3. J. Biochem. Mol. Biol. 39, 537–545 (2006)
F.Z. Hu, Z.Z. Sheng, C.P. Qin, T. Xu, Research advances in CKLFSF-like MARVEL transmembrane domain containing member 3. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 38, 360–363 (2016)
S. Delic, A. Thuy, M. Schulze, M.A. Proescholdt, P. Dietrich, A.K. Bosserhoff, M.J. Riemenschneider, Systematic investigation of CMTM family genes suggests relevance to glioblastoma pathogenesis and CMTM1 and CMTM3 as priority targets. Genes Chromosomes Cancer 54, 433–443 (2015)
Z. Zhou, Z. Ma, Z. Li, H. Zhuang, C. Liu, Y. Gong, S. Huang, C. Zhang, B. Hou, CMTM3 overexpression predicts poor survival and promotes proliferation and migration in pancreatic cancer. J. Cancer. 12, 5797–5806 (2021)
S. Li, P. Gao, X. Dai, L. Ye, Z. Wang, H. Cheng, New prognostic biomarker CMTM3 in low grade glioma and its immune infiltration. Ann. Transl. Med. 10, 206 (2022)
W. Yuan, T. Li, X. Mo, X. Wang, B. Liu, W. Wang, Y. Su, L. Xu, W. Han, Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget 7, 29507–29519 (2016)
W. Yuan, B. Liu, X. Wang, T. Li, H. Xue, X. Mo, S. Yang, S. Ding, W. Han, CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 386, 77–86 (2017)
W. Yuan, F. Wei, H. Ouyang, X. Ren, J. Hang, X. Mo, Z. Liu, CMTM3 suppresses chordoma progress through EGFR/STAT3 regulated EMT and TP53 signaling pathway. Cancer Cell Int. 21, 510 (2021)
W. Li, S. Zhang, CKLF-Like MARVEL transmembrane domain-containing member 3 (CMTM3) inhibits the proliferation and Tumorigenisis in hepatocellular carcinoma cells. Oncol. Res. 25, 285–293 (2017)
Z.L. Yu, Z.M. Zhu, Comprehensive analysis of N6-methyladenosine -related long non-coding RNAs and immune cell infiltration in hepatocellular carcinoma. Bioengineered 12, 1708–1724 (2021)
N.F. Lange, V. Graf, C. Caussy, J.F. Dufour, PPAR-targeted therapies in the treatment of non-alcoholic fatty liver disease in diabetic patients. Int. J. Mol. Sci. 23, (2022)
T. Chi, M. Wang, X. Wang, K. Yang, F. Xie, Z. Liao, P. Wei, PPAR-gamma modulators as current and potential cancer treatments. Front. Oncol. 11, 737776 (2021)
J. Yu, L. Qiao, L. Zimmermann, M.P. Ebert, H. Zhang, W. Lin, C. Rocken, P. Malfertheiner, G.C. Farrell, Troglitazone inhibits tumor growth in hepatocellular carcinoma in vitro and in vivo. Hepatology 43, 134–143 (2006)
J. Cheng, H. Nakamura, H. Imanishi, W. Liu, T. Morisaki, T. Sugiyama, T. Hada, Peroxisome proliferator-activated receptor gamma ligands, 15-deoxy-Delta 12,14-prostaglandin J2, and ciglitazone, induce growth inhibition and cell cycle arrest in hepatic oval cells. Biochem. Biophys. Res. Commun. 322, 458–464 (2004)
K.R. Kim, H.N. Choi, H.J. Lee, H.A. Baek, H.S. Park, K.Y. Jang, M.J. Chung, W.S. Moon, A peroxisome proliferator-activated receptor gamma antagonist induces vimentin cleavage and inhibits invasion in high-grade hepatocellular carcinoma. Oncol. Rep. 18, 825–832 (2007)
Y.I. Liu, Z. Liu, Y. Chen, K. Xu, J. Dong, PPARgamma activation reduces ischemia/reperfusion-induced metastasis in a murine model of hepatocellular carcinoma. Exp. Ther. Med. 11, 387–396 (2016)
X. Zhou, Y. Chi, Z. Dong, T. Tao, X. Zhang, W. Pan, Y. Wang, A nomogram combining PPARgamma expression profiles and clinical factors predicts survival in patients with hepatocellular carcinoma. Oncol. Lett. 21, 319 (2021)
Y. Shu, Y. Lu, X. Pang, W. Zheng, Y. Huang, J. Li, J. Ji, C. Zhang, P. Shen, Phosphorylation of PPARgamma at Ser84 promotes glycolysis and cell proliferation in hepatocellular carcinoma by targeting PFKFB4. Oncotarget 7, 76984–76994 (2016)
F. Liu, X. Liu, X. Liu, T. Li, P. Zhu, Z. Liu, H. Xue, W. Wang, X. Yang, J. Liu, W. Han, Integrated analyses of phenotype and quantitative proteome of CMTM4 deficient mice reveal its association with male fertility. Mol. Cell. Proteomics. 18, 1070–1084 (2019)
J. Wang, X. Liu, H.J. Chu, N. Li, L.Y. Huang, J. Chen, Centromere Protein I (CENP-I) is upregulated in gastric cancer, predicts poor prognosis, and promotes tumor cell proliferation and migration. Technol Cancer Res. Treat. 20, 15330338211045510 (2021)
D. Kim, B. Langmead, S.L. Salzberg, HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015)
Expression of PPARG in liver cancer, in The Human Protein Atlas [Internet]. https://www.proteinatlas.org/ENSG00000132170-PPARG/pathology/liver+cancer Accessed 1 May 2022
D.J. Pritchard, W.H. Butler, Apoptosis–the mechanism of cell death in dimethylnitrosamine-induced hepatotoxicity. J. Pathol. 158, 253–260 (1989)
G. Jayakumar Amirtharaj, K.R. Thangaraj, A. Kini, V. Raghupathy, A. Goel, C.E. Eapen, A. Venkatraman, A.B. Pulimood, A.K. Balasubramanian, A. Ramachandran, Acute liver injury induced by low dose dimethylnitrosamine alters mediators of hepatic vascular flow. Toxicol. Rep. 1, 707–717 (2014)
H. Robles, in Encyclopedia of Toxicology, ed. by P. Wexler (Academic Press, 2014), p. 584–585
R. Tolba, T. Kraus, C. Liedtke, M. Schwarz, R. Weiskirchen, Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab. Anim. 49, 59–69 (2015)
I. Syed, J. Rathod, M. Parmar, G.B. Corcoran, S.D. Ray, Matrix metalloproteinase-9, -10, and -12, MDM2 and p53 expression in mouse liver during dimethylnitrosamine-induced oxidative stress and genomic injury. Mol. Cell. Biochem. 365, 351–361 (2012)
D.J. Lees Murdock, Y.A. Barnett, C.R. Barnett, DNA damage and cytotoxicity in pancreatic beta-cells expressing human CYP2E1. Biochem. Pharmacol. 68, 523–530 (2004)
D.L. van der Meer, T. Degenhardt, S. Vaisanen, P.J. de Groot, M. Heinaniemi, S.C. de Vries, M. Muller, C. Carlberg, S. Kersten, Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 38, 2839–2850 (2010)
J. George, G. Chandrakasan, Biochemical abnormalities during the progression of hepatic fibrosis induced by dimethylnitrosamine. Clin. Biochem. 33, 563–570 (2000)
Z. Bahadoran, A. Ghasemi, P. Mirmiran, F. Azizi, F. Hadaegh, Nitrate-nitrite-nitrosamines exposure and the risk of type 1 diabetes: A review of current data. World J. Diabetes 7, 433–440 (2016)
I. Chrifi, L. Louzao-Martinez, M. Brandt, C.G.M. van Dijk, P. Burgisser, C. Zhu, J.M. Kros, D.J. Duncker, C. Cheng, CMTM3 (CKLF-Like Marvel Transmembrane Domain 3) mediates angiogenesis by regulating cell surface availability of VE-Cadherin in endothelial adherens junctions. Arterioscler. Thromb. Vasc. Biol. 37, 1098–1114 (2017)
T. Yamauchi, J. Kamon, H. Waki, Y. Terauchi, N. Kubota, K. Hara, Y. Mori, T. Ide, K. Murakami, N. Tsuboyama-Kasaoka, O. Ezaki, Y. Akanuma, O. Gavrilova, C. Vinson, M.L. Reitman, H. Kagechika, K. Shudo, M. Yoda, Y. Nakano, K. Tobe, R. Nagai, S. Kimura, M. Tomita, P. Froguel, T. Kadowaki, The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 7, 941–946 (2001)
N. Stefan, M. Stumvoll, Adiponectin–its role in metabolism and beyond. Horm. Metab. Res. 34, 469–474 (2002)
M. Fer, L. Corcos, Y. Dreano, E. Plee-Gautier, J.P. Salaun, F. Berthou, Y. Amet, Cytochromes P450 from family 4 are the main omega hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. J. Lipid Res. 49, 2379–2389 (2008)
T. Han, Y. Lv, S. Wang, T. Hu, H. Hong, Z. Fu, PPARgamma overexpression regulates cholesterol metabolism in human L02 hepatocytes. J. Pharmacol. Sci. 139, 1–8 (2019)
E. Gonzalez, T.E. McGraw, The Akt kinases: isoform specificity in metabolism and cancer. Cell Cycle 8, 2502–2508 (2009)
J. Sanchez-Gurmaches, C. Martinez Calejman, S.M. Jung, H. Li, D.A. Guertin, Brown fat organogenesis and maintenance requires AKT1 and AKT2. Mol. Metab. 23, 60–74 (2019)
Acknowledgements
We thank Prof. Wenling Han from Peking University Center for Human Disease Genomics, China for generously providing resources and expertise, Prof. Jian Chen from the Medical Oncology Department, Yantai Yuhuangding Hospital for providing human tissue samples, and Mr. Jun Li of the Central Laboratory, Yantai Yuhuangding Hospital for animal maintenance.
Funding
This work was supported by grants from the Shandong Medicine and Health Science and Technology Development Plan, China (Grant no.: 202102080625, 202102080643), National Natural Science Foundation of China (Grant no.: 81971438), and Natural Science Foundation of Shandong Province (Grant no.: ZR2020MH075). The funding body was not involved in any form in the design of the study and collection, analysis, interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
F.L. and conceived of this study, supervised the project and critically revised the manuscript. J.W. primarily performed the investigations, analyzed data, and wrote the first draft manuscript. H.C., Z.W., X.L., Z.S., and X.W. participated in material preparation, data collection and analysis. All authors commented on previous versions of the manuscript, and have read and approved the final manuscript.
Corresponding author
Ethics declarations
Research involving human participants and/or animals
This study was approved by the Ethics Committee of Yantai Yuhuangding Hospital (IRB: 2019–398). All procedures for animal care and use were carried out following the guidelines for the care and use of laboratory animals in accordance with the US NIH Guide for the Care and Use of Laboratory Animals (no. 8023, revised in 1996). All efforts were made to reduce the number of animals used and bodily suffering during the experiment.
Informed consent
Written informed consent was obtained from all participants for the use of human samples and clinical information for research and publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Chu, H., Wang, Z. et al. In vivo study revealed pro-tumorigenic effect of CMTM3 in hepatocellular carcinoma involving the regulation of peroxisome proliferator-activated receptor gamma (PPARγ). Cell Oncol. 46, 49–64 (2023). https://doi.org/10.1007/s13402-022-00733-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-022-00733-1