Abstract
Peptide cation radicals of the z-type were produced by electron transfer dissociation (ETD) of peptide dications and studied by UV-Vis photodissociation (UVPD) action spectroscopy. Cation radicals containing the Asp (D), Asn (N), Glu (E), and Gln (Q) residues were found to spontaneously isomerize by hydrogen atom migrations upon ETD. Canonical N-terminal [z4 + H]+● fragment ion-radicals of the R-C●H-CONH- type, initially formed by N−Cα bond cleavage, were found to be minor components of the stable ion fraction. Vibronically broadened UV-Vis absorption spectra were calculated by time-dependent density functional theory for several [●DAAR + H]+ isomers and used to assign structures to the action spectra. The potential energy surface of [●DAAR + H]+ isomers was mapped by ab initio and density functional theory calculations that revealed multiple isomerization pathways by hydrogen atom migrations. The transition-state energies for the isomerizations were found to be lower than the dissociation thresholds, accounting for the isomerization in non-dissociating ions. The facile isomerization in [●XAAR + H]+ ions (X = D, N, E, and Q) was attributed to low-energy intermediates having the radical defect in the side chain that can promote hydrogen migration along backbone Cα positions. A similar side-chain mediated mechanism is suggested for the facile intermolecular hydrogen migration between the c- and [z + H]●-ETD fragments containing Asp, Asn, Glu, and Gln residues.

ᅟ
Similar content being viewed by others
Introduction
Electron transfer dissociation (ETD) of multiply charged peptide and protein ions proceeds by homolytic dissociation of bonds between the amide nitrogen and Cα position of the adjacent amino acid residues, N−Cα bond cleavage for short [1, 2]. N−Cα bond cleavage is orthogonally complementary to the heterolytic and proton driven cleavage of peptide amide CO-NH bonds [3]. N−Cα bond cleavage produces N-terminal even-electron fragment ions of the [cm+2H]+ type and C-terminal odd-electron fragment ions of the [zn+H]+● type, where m and n are the respective numbers of amino acid residues in the fragment ions, that provide important information on the peptide sequence [4, 5]. For an all-inclusive nomenclature of peptide radical ions see ref [6]. The currently accepted Utah-Washington mechanism of ETD [7,8,9] describes the formation of the [cm+2H]+ and [zn+H]+● fragment ions as sketched in Scheme 1. The [cm+2H]+ ions can be formed with C-terminal enolimine groups or as the substantially more stable amide isomers [10]. The [zn+H]+● fragment ions are usually presented as N-terminally deaminated radicals, R-C●H-CONH-, with the charge being carried by a protonated basic residue (Lys or Arg). We call such structures the canonical [zn+H]+● ions. The actual structures of a few [cm+2H]+ and [zn+H]+● fragment ions formed by electron-based dissociation have been studied by action spectroscopy [11,12,13,14,15,16]. This technique is based on resonant absorption of one or multiple photons over a range of wavelengths, which leads to dissociation and formation of fragment ions. A wavelength-dependent profile of the fragment ion intensities can be produced and used to track absorbance of the ion under study for comparison with its calculated absorption spectrum [17, 18]. The few [cm + 2H]+ ions studied so far by infrared multiphoton dissociation action spectroscopy (IRMPD) have been found to have C-terminal amide groups [11, 12]. This is consistent with computational studies that indicated very low activation energies for proton-enabled enol imine-amide isomerizations in {[cm+2H]…[zn+H]}+● complexes formed by N−Cα bond cleavage [19,20,21]. In contrast, [zn+H]+● ion structures appear to depend on the constituent amino acid residues. For example, the authors of an IRMPD study of [zn+H]+● ions (n = 1–4) produced by ETD of (AAHAR+2H)2+ concluded that the ions had canonical, N-terminally deaminated radical structures [13]. In contrast, a UVPD study of [z4+H]+● ions, corresponding to [●AHAR+H]+, has found ca. 6% of non-canonical isomers [14]. Further amino acid residue effects have been reported for [z4+H]+● ions corresponding to [●AWAR+H]+ and [●AFAR+H]+ for which UVPD action spectroscopy indicated the presence of non-canonical isomers [15]. The non-canonical fractions were composed of respective Trp and Phe β-side chain radicals of the benzylic type, and backbone Cα radicals at the inner residues, although the contributions of the individual isomers have not been quantified [15]. In a yet contrasting result, UVPD studies of [●ASAR+H]+ and [●ATAR+H]+ ions led the authors to conclude that these ions chiefly consisted of canonical structures [16, 22]. A survey UVPD study of a library of [●AXAR + H]+, [●AXAK + H]+, and [●XAR + H]+ ions pointed out the tendency of the Phe and Asp residues to promote ion isomerization upon ETD [14]. These results have indicated that the degree to which canonical structures isomerize depends on the amino acid residues, both those initially carrying the Cα radical and those in adjacent positions.
Hydrogen atom migrations between complementary ETD fragments are known to produce radical [cm+H]+● and even-electron [zn+2H]+ ions [23] that can complicate sequence assignment for residues differing by 1 Da (Asn/Asp and Gln/Glu). In a survey study, Savitski et al. analyzed the relative [zn+2H]+ and [zn+H]+● fragment ion intensities in electron capture dissociation mass spectra of tryptic peptides [24]. Statistical analysis indicated the propensity of Gly-terminated [zn+H]+● ions to abstract a hydrogen atom from their [cm+H] neutral or [cm+2H]+ charged counterparts. Studies [25, 26] of ETD of specific nona- and decapeptides identified the Asp and Asn residues as most inductive for hydrogen transfer accompanying N−Cα bond cleavage between Asp-Asp, Asn-Asn, Asp-Asn, and Asn-Asp residues [26]. The Glu and Gln residues were less effective, and the [zn+2H]+ ion formation rapidly diminished in backbone fragmentations occurring farther away from the Asn, Asp, Gln, and Glu residues [26]. Computational studies [19,20,21] of {[cm+2H]…[zn+H]}+● complexes formed by N−Cα bond cleavage indicated low energy barriers for inter-fragment hydrogen atom transfer that were lower than or comparable to the energy barriers to intramolecular H-atom migrations in peptide cation radicals, as studied for several systems [27,28,29,30,31,32]. Thus, it was of interest to study structures of [zn+H]+● fragment ions containing Asp, Asn, Glu, and Gln residues to elucidate possible connections between inter-fragment and intra-molecular H-atom migrations triggered spontaneously upon ETD.
Experimental
Materials and Methods
Peptides QDAAR and QNAAR were custom made at >90% purity by GenScript (Piscataway, NJ, USA). Peptides QQAAR and QEAAR were synthesized on Wang resin using standard solid-phase methods. The sequences of all peptides were checked by electrospray ionization mass spectra. The ETD mass spectra were measured on a ThermoElectron Fisher LTQ-XL-ETD linear ion trap mass spectrometer with a fluoranthene anion reagent and ion reaction times of 100–300 ms. UVPD action spectra were measured as described previously [33]. Briefly, the [z4+H]+● ions were generated by ETD in the ion trap, selected by mass and exposed to laser pulses (3–6 ns pulse width, 20 Hz repetition rate) generated by an EKSPLA NL301G (Altos Photonics, Bozeman, MT, USA) Nd:YAG laser source equipped with a PG142C optical parametric oscillator. This provided wavelength tuning in the range of 210–700 nm with a laser power of 0.52–12.69 mJ per pulse. The intensities of the resulting UVPD MS3 fragment ions were monitored as a function of wavelength, normalized to the laser output power at each wavelength, and plotted to provide action spectra.
Calculations
Cation-radical structures were obtained at several levels of theory, starting with conformer analysis and followed by full gradient geometry optimization, transition state search, and intrinsic reaction coordinate analysis [34]. Conformer analysis was performed by Born-Oppenheimer Molecular Dynamics (BOMD) calculations using the semi-empirical all-electron PM6 method [35] supplemented with dispersion corrections [36], PM6-D3H4, and run by MOPAC [37] under the Cuby4 platform [38], as described previously [16]. Selected low-energy conformers from the BOMD runs were re-optimized by density functional theory (DFT) calculations in a spin-unrestricted formalism using the B3LYP [39], ωB97X-D [40], and M06-2X [41] hybrid functionals with the 6-31+G(d,p) basis set. The structures were confirmed as local energy minima by harmonic frequency analysis. Energy ranking was based on single-point energy calculations with DFT and Møller-Plesset [42] (UMP2, frozen core) using the 6-311++G(2d,p) basis set. Higher spin states in UMP2 energies were annihilated by spin projection [43, 44], providing spin expectation values of <0.78 for local energy minima and <0.9 for transition states. Additionally, restricted open-shell single point MP2 calculations (ROMP/6-311++G(2d,p)) [45] were performed to check the effect of spin contamination [46]. Excitation energies and oscillator strengths were obtained for 25 excited states by time-dependent DFT calculations [47] with ωB97X-D and M06-2X/6-31+G(d,p), according to a previous benchmarking study [48]. Vibronically broadened absorption spectra were obtained by multiple TD-DFT calculations of 16 excited states over 200–300 random ground state vibrational configurations generated by Newton-Xspace software (ver. 2.0, www.newtonx.org) [49, 50] and weighted according to their Boltzmann factors at 310 K. All the electronic structure calculations were performed with the Gaussian 09 (Revision A.02) suite of programs [51]. Rice-Ramsperger-Kassel-Marcus (RRKM) calculations [52] of unimolecular rate constants were performed using the QCPE program [53] that was recompiled for Windows [54]. The RRKM calculations were run with direct count of quantum states and molecular rotations were treated adiabatically. The k(E, J, K) rate constants were Boltzmann averaged over the rotational state distribution at 300 K.
Results and Discussion
Ion Formation and Photodissociation
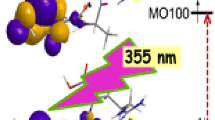
ETD of doubly charged (XDAAR+2H)2+ (X = A, Q), (QNAAR+2H)2+, (QQAAR+2H)2+, and (QEAAR+2H)2+ peptide ions produced the respective [zn+H]+● ions (Supplementary Figure S1a–d, Supporting Information). The [z4+H]+● ions from each sequence were selected by mass and further analyzed by CID and UVPD-MS3 spectra, as illustrated for [●DAAR+H]+ (Figure 1a, b). The CID-MS3 spectrum of the [●DAAR+H]+ ion at m/z 416 displayed a dominant fragment ion by loss of CO2 that was accompanied by a [z3+H]+● ion at m/z 301 (Figure 1a). In contrast, UVPD-MS3 spectrum at 355 nm resulted in a backbone amide cleavage at the Asp residue, forming the [y3]+ fragment ion at m/z 315. Another amide backbone cleavage produced the [y2]+ fragment ion at m/z 244 (Figure 1b). Analogous backbone dissociations have been previously observed for UVPD of other [zn+H]+● ions [14,15,16, 22]. The pulse dependence experiments at 355 nm (Figure 1c) showed a complete depletion of the [●DAAR+H]+ ions fitting the single-exponential decay equation: I(n) = I0e−αn, where I(n) is the residual ion intensity after n pulses normalized to the initial ion intensity I0, and α = 0.537 is the photodissociation efficiency at 355 nm. The fit gave a root-mean square-deviation of 1.3%. The profiles of the UVPD fragment ions showed an asymptotic increase of the [y2]+ and [y3]+ ion intensities at a constant ratio of [y2]+/[y3]+ = 0.29 ± 0.03. These are even-electron ions that do not absorb light at 355 nm. The cation-radical fragments at m/z 372 (loss of CO2) and [z3+H]+● (m/z 301) showed an initial increase followed by depletion, which was complete for the latter ion after ca. 10 laser pulses. In contrast, a fraction of the m/z 372 ions persisted even after 10 laser pulses. The complete exponential decrease of the [●DAAR+H]+ ion intensity indicated that the ion population produced by ETD consisted of species that absorbed light at 355 nm, whereas non-absorbing isomers were absent or very weak (<1%). We note that this result is different from photodissociation of the homologous [●DAR+H]+ ions that revealed 41% of residual non-dissociating species [14].
UVPD Action Spectra
The [z4+H]+● ions were further characterized by photodissociation action spectra in the 210–700 nm region. The action spectrum of [●DAAR+H]+ showed several bands in the mass-resolved dissociation channels with maxima at 212, 270, 335–350, and 395 nm (Figure 2). The signal of the [y2]+ fragment ion was compromised by photodesorbed ions from the ETD reagent fluoranthene [33] that became prominent at shorter wavelengths and therefore was not plotted in Figure 2. No photodissociation was observed above 500 nm. The main channel producing the [y3]+ fragment ion was prominent in the 210–370 nm region. Note that the even-electron [y3]+ ion is not expected to undergo photodepletion in this region. In contrast, the profile of the [z3+H]+● fragment ion is affected by photodissociation at 355 nm (Figure 1c). The 395 nm band was represented in the channels for the CO2 loss and [z3+H]+● fragment ions. The CO2 loss ion disappeared below 300 nm, possibly because of subsequent photodissociation. This was ascertained by obtaining the action spectrum of the m/z 372 fragment ion formed by CO2 loss from [●DAAR+H]+ upon CID-MS4 (Supplementary Figure S2). The action spectrum of the m/z 372 fragment ion showed absorption bands at 330, 270, and 230 nm, consistent with the photodepletion of this channel in the 210–300 nm region of the [●DAAR+H]+ precursor action spectrum.
The action spectrum of [●NAAR+H]+ (Figure 3a) differed from that of [●DAAR+H]+ in that it did not show strong absorption bands above 360 nm. The main bands were represented by the m/z 371 fragment ion (assigned to a loss of CONH2 radical from Asn) that showed broad bands with maxima at 240 and 275 nm (Figure 3a). Loss of CONH2 was also prominent in the ETD-CID-MS3 spectrum of [●NAAR+H]+ (Supplementary Figure S3a). The [y3]+ ion channel (m/z 315) was weaker in the UVPD spectrum, showing absorption maxima at 230 and 275 nm (Figure 3a). The [z3+H]+● channel (m/z 301) showed a broad band with a maximum at 260 nm.
The action spectra of [●EAAR+H]+ and [●QAAR+H]+ showed some similarities (Figure 3b, c). In both spectra, the dominant photodissociation channel was at m/z 371 for loss of 59 Da and 58 Da neutral fragments, corresponding to the CH2COOH and CH2CONH2 radicals from the Glu and Gln side chains, respectively (Figure 3b, c). These dissociations were also prominent in the ETD-CID-MS3 spectra of the corresponding [z4+H]+● ions (Supplementary Figure S3b, c). The backbone [y3]+ fragment ions at m/z 315 were only minor upon UVPD. Both spectra showed weak absorption bands at 390–400 nm and broad bands with maxima at 340–350 and 270 nm. The main absorption bands were at 230 and 220 nm for [●EAAR+H]+ and [●QAAR+H]+, respectively.
Excited-State Calculations of UV-Vis Absorption Spectra
Interpretation of the UV-Vis action spectra of gas-phase peptide ions relies on calculations providing electronic excitation energies and oscillator strengths of several valence bond isomers, each being represented by multiple low-energy conformers. This generates vibration-less (0 K) absorption spectra for each species that are further expanded by accounting for transitions starting from multiple vibrational states of the ion ground electronic state at the temperature of the action spectroscopy measurement. We applied this combined approach to analyze the action spectrum of [●DAAR+H]+. Conformational analysis of canonical [●DAAR+H]+ ions produced low-energy conformers that are represented by structures 1a-1f (Figure 4, Table 1). All these low-energy structures showed hydrogen bonding between the charged Arg guanidine group and the Asp amide carbonyl. The narrow range of free energies within this group, along with the estimated ±10 kJ mol-1 inaccuracy of the DFT relative energy calculations, indicated that any of these structures could be present under equilibrium conditions in the gas phase. Structure 1b was the lowest free-energy isomer by M06-2X and ωB97X-D calculations (Table 1) and was therefore selected as a reference in further calculations (vide infra).
The ωB97X-D TD-DFT spectra of 1a-1f showed an electronic transition at 330–371 nm corresponding to the second (B) excited state (Supplementary Figure S4). This state arises by electron excitation from an underlying doubly occupied Asp amide πz molecular orbital (MO110) to the semi-occupied πz molecular orbital at the -Cα●H-CONH group (MO111) of the same symmetry. Analogous excitations have been found for other peptide canonical [z4+H]+● ions [16, 22]. The πz → πz excitation energy was sensitive to the ion conformation affecting the geometry at the Asp Cα atom (Supplementary Figure S4). Conformers 1a-1c, 1e, and 1f in which the -C●H-CO group was distorted by 4–11 degrees out of the plane at the Cα atom showed the B-state transition below 350 nm. This puckering at Cα was forced by hydrogen bonding of the Asp COOH and amide groups to the Arg guanidinium (Figure 4). In contrast, conformer 1d had nearly planar -C●H-CO group that resulted in a red shift of the B state excitation energy to 371 nm. Accordingly, the dipole disfavored πxy → πz excitation corresponding to the first (A) excited state at 406 nm had a zero intensity in 1d, whereas it was apparent in the other conformers with a higher degree of out-of-plane distortion, e.g., the 366 nm line for 1b (Supplementary Figure S4 and Figure 5a). The vibronically broadened absorption spectra of 1b and 1d (Figure 5a, b) showed bands in the 300–450 nm region that weakly appeared in the same region of the action spectrum. In addition, a prominent narrow band at 230 nm in the calculated spectra had a matching counterpart at 220 nm in the action spectrum. However, a conspicuous feature of the absorption spectra of 1a-1f was the absence of a strong band in the 270–320 nm region whereby the action spectrum showed strong absorption. To account for these bands, it was necessary to consider [●DAAR+H]+ isomers other than the canonical ions 1a-1f. The calculated vibronically broadened spectra of several Cα and Cβ radical isomers are shown in Figure 5a–f.
Vibronically broadened (310 K) absorption spectra from Greek omega B97X-D/6-31+G(d,p) TD-DFT calculations of (a) 1b, (b) 1d, (c) 2, (d) 3, (e) 4, and (f) 5. The black bars represent the calculated electronic transitions at 0 K. The green lines are error bars from Newton-X vibronic transition calculations at 310 K
Structure 2 is an isomer derived from 1 by a 1,2-H shift moving the radical site to the β-position of the Asp side chain next to the COOH group (Figure 5c). The vibronically broadened TD-DFT absorption spectrum of 2 showed bands with maxima at 260 and 300 nm and a shoulder extending to 350 nm. The spectrum of the Ala-2 Cα radical isomer 3 showed several bands peaking at 255 and 285 nm and extending as a broad shoulder to 400 nm (Figure 5d). Another isomer showing absorption in this region was the Ala-3 Cα radical 4 (Figure 5e) showing intense absorption bands with maxima at 240 and 265 nm and a weaker band at 325 nm. Finally, the Arg-4 Cα-radical 5 displayed several bands in the 245–300 nm region that were vibronically broadened to longer wavelengths (Figure 5f).
A comparison of the action spectrum with the calculated absorption spectra of 1-5 indicates that the [●DAAR+H]+ ions produced by ETD did not consist of a single species but comprised a mixture of radical isomers. Evidence for a conformer of 1 is provided by the 220 and 400 nm bands that are unique to the canonical ion structure. The other isomers showed overlapping vibronic spectra in the 250–350 nm region and could not be distinguished in the action spectrum taken at ambient temperature. Of note is that the absorption spectrum of radical 2 displayed a reasonable fit to the 250–350 nm region of the action spectrum. However, the pertinent bands for 2 had low oscillator strengths, and so this isomer could contribute to the action spectrum only if it was a major component.
Potential Energy Surface for [DAAR+H]+●
The spontaneous formation upon ETD of [z4+H]+● isomers other than the canonical structure (1) raised the question of the energetics and kinetics of unimolecular isomerizations of 1. These can occur in vibrationally excited [z4+H]+● ions in a unimolecular fashion after fragment separation [55] or in a [{[cm+2H]…[zn+H]}+● complex following N–Cα bond cleavage. Here we focus on unimolecular isomerizations. Using calculations at several levels of theory (Table 2), we obtained the relevant parts of the potential energy surface for 1, several of its isomers, interconnecting transition states (TS), and dissociation products. The optimized intermediate and TS structures are compiled in Supplementary Schemes S1–S5 of the Supporting Information. The potential energy surface, based on combined B3LYP and PMP2/6-311++G(2d,p) calculations, referred to as B3-PMP2, [56, 57], is plotted in Figure 6. The lowest TS energy for isomerization of 1 pertains to an Asp amide trans-cis rotation (TS1) forming cis-1, followed by transfer of the Ala-2-Hα via TS2 forming cis-3 (Scheme 2). The cis-Ala-2 Cα radical can readily stabilize by amide rotation through a low-lying TS3 to form the trans isomer 3. Competitively, cis-3 can undergo isomerization by Asp β-H migration (TS4), forming cis-2, which can stabilize by amide rotation via TS5 to the trans amide isomer 2. This seemingly complicated pathway to 2 is energetically more favorable than a direct 1,2-H migration in 1 via TS6 (Figure 6, Supplementary Scheme S3). Significantly, all these isomerization TS energies are lower than the TS or threshold energies for dissociations. This concerns the CO–NH bond cleavage via TS7 and complex 6 to form the [y3]+ ion and the complementary HOOCCH2CH2CO● neutral fragment (Supplementary Scheme S4). Likewise, a Cα−CO bond dissociation in 2 (TS8), proceeding to a complex (7) and further to [x3+H]+● and [z3+H]+● fragment ions, requires a threshold energy that is above the TS for isomerizations (Supplementary Scheme S5). A relatively high-energy transition state (TS9) and dissociation threshold also apply to the loss of COOH from 1, forming ion 8 (Supplementary Scheme S6). Rather unexpectedly, the carboxyl hydrogen migration to Asp Cα was calculated to have a relatively high TS energy (TS10) for isomerization to COO radical 9, followed by loss of CO2 via TS11 and forming ion 10 (Supplementary Scheme S6). The high energy for TS10 contradicted the very facile loss of CO2 upon CID (Figure 1a). We did not investigate this point in more detail, although we note that Chu and coworkers obtained a similar B3LYP energy for a TS of an analogous Asp carboxyl hydrogen atom migration in a tripeptide cation radical [32]. Overall, the loss of CO2 is exothermic (Table 2) and thus irreversible, which may contribute to its kinetic prominence compared with competing dissociations involving stable ion-molecules complexes 6 and 7. The unambiguous conclusion of the PES analysis is that 1, 2, and 3 can undergo isomerizations at internal energies that do not result in dissociation. Furthermore, the 1, 2, and 3 energies are very close on the PES, indicating these isomers can be comparably populated if an isomerization equilibrium was achieved.
This assumption was supported by RRKM kinetic analysis of the pertinent reaction steps in Scheme 2. Isomerization of 1 via TS1, cis-1, and TS2 is illustrated by the conversion of 1, which represents the fraction that passed through TS2 at reaction times ranging from 0.01 to 0.05 s (Figure 7a). These reaction times were considered with regard to fast collisional cooling in the ion trap that proceeds on a similar time scale [55]. The internal energies needed to achieve 50% conversion of 1 ranged from 322 to 359 kJ mol-1 for the 0.05 and 0.01 s reaction times, respectively. This is to be compared with the TS2 energy shown by an arrow in Figure 7a (111 kJ mol-1, Table 2), indicating a substantial kinetic shift for the isomerization. The ion energies needed to achieve kinetically relevant isomerization are compared with the estimated excitation energy of 1 produced by ETD from (QDAAR+2H)2+. In making the estimate, we included the thermal distribution of vibrational enthalpy in the precursor ion (QDAAR+2H)2+ at the ion trap temperature of 310 K, exothermicity of electron transfer from fluoranthene, ΔE = RE[(QDAAR+2H)2+] – EA(fluoranthene) = 434 – 58 = 376 kJ mol-1, and energy partitioning between the c1+H and [z4+H]+ fragments according to their heat capacities [55, 58]. This yielded the mean vibrational energy of 1, Emean = 382 kJ mol-1, with a distribution shown in Figure 7b. The estimated internal energy distribution in 1 fits the kinetically relevant energy interval for isomerization (Figure 7a) and is consistent with the experimental data showing spontaneous isomerization upon ETD.
The product distribution among 1, 2, and 3 was treated using a simplified kinetic scheme, 1 → TS2 → 3 → TS4 → 2, where all steps were treated as reversible reactions with pertinent RRKM rate constants. Figure 8 shows the energy-dependent molar fractions of 1, 2, and 3, indicating 2 as a major isomerization product, but also revealing a substantial reverse isomerization to 1. Note that the order of ion potential energies was 1> 2 > 3, making 1 marginally least stable according to the ΔH0 values. The finding that 1 was kinetically preferred can be attributed to its high vibrational entropy and density of vibrational states that lowered the rate constant for the 1 → TS2 → 3 isomerization relative to the reverse reaction. It should be noted that we did not investigate further isomerizations of 2 to the more remote Cα positions of Ala-3 (4) and Arg-4 (5). However, both these radicals are thermodynamically more stable than 1-3 (Table 2) and, if kinetically accessible, would represent cul-de-sac traps of the non-dissociating isomer group.
[NAAR+H]+●, [EAAR+H]+●, and [QAAR+H]+● Isomers
The previous analysis of the [DAAR+H]+● action spectrum can be extended to the other sequences with the caveat that we did not have detailed theoretical absorption spectra for all isomers of these ions. The conserved feature in the absorption spectra of the canonical [XAAR+H]+● ions [15, 16] is the prominent bands at 220 and >350 nm and lack of absorption between 250 and 330 nm. The action spectrum of [NAAR+H]+● (Figure 3a) showed a prominent band at 275 nm that indicated the presence of non-canonical isomer(s) formed by spontaneous isomerization upon ETD. An analogous band at 270 nm also appeared in the action spectra of [QAAR+H]+● and [EAAR+H]+●, (Figure 3b and c, respectively) albeit at a lower intensity than for [NAAR+H]+● and [DAAR+H]+●. This allowed us to conclude that these [XAAR+H]+● ions (X = Asn, Glu, and Gln) also underwent partial isomerization upon ETD.
Internal Versus Inter-Fragment Hydrogen Migration in [zn+H]+● Ions and Ion-Molecule Complexes
The high propensity of the Asp residue to promote spontaneous intramolecular isomerization of [DAAR+H]+● ions can be related to the facile hydrogen migration to Asp-terminated [zn+H]+● fragment ions accompanying backbone cleavage in ETD [25, 26]. Migration of a Hα from a c-type amide fragment to a [zn+H]+● radical in an ion-molecule complex is exothermic and faces a low energy barrier [20] that was calculated to depend on the ion charge state [19, 21]. The range of calculated TS energies is comparable to the typical binding energies of the [c + z]+● complexes [2, 59, 60], making the hydrogen transfer energetically competitive. However, the hydrogen transfer kinetics is affected by the entropically disfavored reorientation of the reactants in the complex to reach the transition state [21]. This can slow down the hydrogen transfer, which is a tight transition state reaction, in competition with fragment separation, which is a continuously endothermic process that can be realized in a multitude of configurations. The Asp effect can be understood as involving a facile isomerization to an Asp β-radical, analogous to 2, whereby the side-chain radical can access a Cα–H on the c-type fragment for exothermic hydrogen transfer. This is essentially a steric effect [19, 21] analogous to the well documented tendency of primary Cα radicals in glycine-N-terminated [zn+H]+● ions to undergo inter-fragment hydrogen transfer [24].. Experimental verification of [zn+H]+● ion isomerization in ion-molecule complexes can potentially be achieved by the following sequence. First, ETD of a suitable peptide dication is used to form charge-reduced cation radicals, presumably {[cm+2H]…[zn+H]}+● ion-molecule complexes. Next, [zn+H]+● ions are produced by CID-MS3 of the complexes and probed by action spectroscopy. Further experimental studies are needed to address this approach and resolve potential difficulties with assigning the charge-reduced cation radical structure [61].
Conclusions
Results from this combined experimental and computational study allow us to arrive at the following conclusions. Asp, Asn, Glu, and Gln residues facilitate hydrogen atom migrations in [zn + H]+● cation-radicals occurring spontaneously upon electron transfer dissociation of peptide dications. The isomerizations produce stable intermediates with the radical defect in the side chain at the alpha position to the carbonyl group. Analogous isomerizations are proposed to facilitate inter-fragment hydrogen atom migrations forming [zn + 2H]+ ions in ETD of peptide ions.
References
Syka, J.E.P., Coon, J.J., Schroeder, M.J., Shabanowitz, J., Hunt, D.F.: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 101, 9528–9533 (2004)
Tureček, F., Julian, R.R.: Peptide radicals and cation radicals in the gas phase. Chem. Rev. 113, 6691–6733 (2013)
Savitski, M.M., Kjeldsen, F., Nielsen, M.L., Zubarev, R.A.: Complementary sequence preferences of electron capture dissociation and vibrational excitation in fragmentation of polypeptide polycations. Angew. Chem. Int. Ed. Engl. 45, 5301–5303 (2006)
Chalkley, R.J., Medzihradszky, K.F., Lynn, A.J., Baker, P.R., Burlingame, A.L.: Statistical analysis of peptide electron transfer dissociation fragmentation mass spectrometry. Anal. Chem. 82, 579–584 (2010)
Li, W., Song, C., Bailey, D.J., Tseng, G.C., Coon, J.J., Wysocki, V.H.: Statistical analysis of electron transfer dissociation pairwise fragmentation patterns. Anal. Chem. 83, 9540–9545 (2011)
Chu, I.K., Siu, C.-K., Lau, J.K.-C., Tang, W.K., Mu, X., Lai, C.K., Guo, X., Wang, X., Li, N., Yao, Z., Xia, Y., Kong, X., Oh, H.-B., Ryzhov, V., Tureček, F., Hopkinson, A.C., Siu, K.W.M.: Proposed nomenclature for peptide ion fragmentation. Int. J. Mass Spectrom. 390, 24–27 (2015)
Sobczyk, M., Anusiewicz, I., Berdys-Kochanska, J., Sawicka, A., Skurski, P., Simons, J.J.: Coulomb-assisted dissociative electron attachment: application to a model peptide. J. Phys. Chem. A. 109, 250–258 (2005)
Syrstad, E.A., Tureček, F.: Toward a general mechanism of electron-capture dissociation. J. Am. Soc. Mass Spectrom. 16, 208–224 (2005)
Chen, X., Tureček, F.: The arginine anomaly: arginine radicals are poor hydrogen atom donors in electron transfer induced dissociations. J. Am. Chem. Soc. 128, 12520–12530 (2006)
Syrstad, E.A., Stephens, D.D., Tureček, F.: Hydrogen atom adducts to the amide bond. Generation and energetics of amide radicals in the gas phase. J. Phys. Chem. A. 107, 115–126 (2003)
Frison, G., Bull, A., van der Rest, G., Tureček, F., Besson, T., Lemaire, J., Maître, P., Chamot-Rooke, J.: Structure of ECD fragments from charge-tagged peptides probed by tunable IRMPD. J. Am. Chem. Soc. 130, 14916–14917 (2008)
Shaffer, C.J., Martens, J., Marek, A., Oomens, J., Tureček, F.: Photoleucine survives backbone cleavage by electron transfer dissociation. A near-UV photodissociation and infrared multiphoton dissociation action spectroscopy study. J. Am. Soc. Mass Spectrom. 27, 1176–1185 (2016)
Martens, J., Grzetic, J., Berden, G., Oomens, J.: Structural identification of electron transfer dissociation products in mass spectrometry using infrared ion spectroscopy. Nat. Commun. 7, 11754 (2016)
Nguyen, H.T.H., Tureček, F.: Near-UV photodissociation of tryptic peptide cation radicals. Scope and effects of amino acid residues and radical sites. J. Am. Soc. Mass Spectrom. 29, 1333–1344 (2017)
Nguyen, H.T.H., Shaffer, C.J., Pepin, R., Tureček, F.: UV action spectroscopy of gas-phase peptide radicals. J. Phys. Chem. Lett. 6, 4722–4727 (2015)
Nguyen, H.T.H., Andrikopoulos, P.C., Bím, D., Rulíšek, L., Dang, A., Tureček, F.: Radical reactions affecting polar groups in threonine peptide ions. J. Phys. Chem. B. 121, 6557–6569 (2017)
Antoine, R., Dugourd, P.: Visible and ultraviolet spectroscopy of gas phase protein ions. Phys. Chem., Chem. Phys. 13, 16494–16509 (2011)
Antoine, R., Dugourd, P.: UV-visible activation of biomolecular ions. (Laser Photodissociation and Spectroscopy of Mass-Separated Biomolecular Ions). Lecture Notes Chem. 83, 93–116 (2013)
Bythell, B.J.: To jump or not to jump? Cα hydrogen atom transfer in post-cleavage radical-cation complexes. J. Phys. Chem. A. 117, 1189–1196 (2013)
Pepin, R., Laszlo, K.J., Peng, B., Marek, A., Bush, M.F., Tureček, F.: Comprehensive analysis of Gly-Leu-Gly-Gly-Lys peptide dication structures and cation-radical dissociations following electron transfer: from electron attachment to backbone cleavage, ion-molecule complexes, and fragment separation. J. Phys. Chem. A. 118, 308–324 (2014)
Bythell, B.J.: Hydrogen atom transfer in post-cleavage radical-cation complexes: short and steep versus long winding road. J. Phys. Chem. A. 118, 10797–10803 (2014)
Nguyen, H.T.H., Shaffer, C.J., Ledvina, A., Coon, J.J., Tureček, F.: Serine effects on collision-induced dissociation and photodissociation of peptide cation radicals of the z+● type. Int. J. Mass Spectrom. 378, 20–30 (2015)
O'Connor, P.B., Lin, C., Cournoyer, J.J., Pittman, J.L., Belyayev, M., Budnik, B.A.: Long-lived electron capture dissociation product ions experience radical migration via hydrogen abstraction. J. Am. Soc. Mass Spectrom. 17, 576–585 (2006)
Savitski, M.M., Kjeldsen, F., Nielsen, M.L., Zubarev, R.A.: Hydrogen rearrangement to and from radical z fragments in electron capture dissociation of peptides. J. Am. Soc. Mass Spectrom. 18, 113–120 (2007)
Chung, T.W., Hui, R., Ledvina, A.R., Coon, J.J., Tureček, F.: Cascade dissociations of peptide cation-radicals. Part 1. Scope and effects of amino acid residues in penta-, nona-, and decapeptides. J. Am. Soc. Mass Spectrom. 23, 1336–1350 (2012)
Ledvina, A.R., Chung, T.W., Hui, R., Coon, J.J., Tureček, F.: Cascade dissociations of peptide cation-radicals. Part 2. Infrared multiphoton dissociation and mechanistic studies of z-ions from pentapeptides. J. Am. Soc. Mass Spectrom. 23, 1351–1363 (2012)
Chung, T.W., Tureček, F.: Backbone and side-chain specific dissociations of z ions from non-tryptic peptides. J. Am. Soc. Mass Spectrom. 21, 1279–1295 (2010)
Hao, Q., Song, T., Ng, D.C.M., Quan, Q., Siu, C.K., Chu, I.K.: Arginine facilitated isomerization: radical-induced dissociation of aliphatic radical cationic glycylarginyl(iso)leucine tripeptides. J. Phys. Chem. B. 116, 7627–7634 (2012)
Zhao, J., Song, T., Xu, M., Quan, Q., Siu, K.W.M., Hopkinson, A.C., Chu, I.K.: Intramolecular hydrogen atom migration along the backbone of cationic and neutral radical tripeptides and subsequent radical-induced dissociations. Phys. Chem., Chem. Phys. 14, 8723–8731 (2012)
Lai, C.-K., Mu, X., Hao, Q., Hopkinson, A.C., Chu, I.K.: Formation, isomerization, and dissociation of ε- and α-carbon-centered tyrosyl glycyl glycine radical cations. Phys. Chem., Chem. Phys. 16, 24235–24243 (2014)
Ledvina, A.R., Coon, J.J., Tureček, F.: Competitive hydrogen atom migrations accompanying cascade dissociations of peptide cation-radicals of the z+● type. Int. J. Mass Spectrom. 377, 44–53 (2015)
Xu, M., Tang, W.-K., Mu, X., Ling, Y., Siu, C.-K., Chu, I.K.: α-Radical induced CO2 loss from the aspartic acid side chain of the collisionally induced tripeptide aspartylglycylarginine radical cation. Int. J. Mass Spectrom. 390, 56–62 (2015)
Shaffer, C.J., Pepin, R., Tureček, F.: Combining UV photodissociation action spectroscopy with electron transfer dissociation for structure analysis of gas-phase peptide cation-radicals. J. Mass Spectrom. 50, 1438–1442 (2015)
Gonzalez, C., Schlegel, H.B.: An improved algorithm for reaction path following. J. Chem. Phys. 90, 2154–2161 (1989)
Stewart, J.J.P.: Optimization of parameters for semi-empirical methods. V. Modification of NDDO approximations and application to 70 elements. J. Mol. Model. 13, 1173–1213 (2007)
Řezáč, J., Fanfrlík, J., Salahub, D., Hobza, P.: Semi-empirical quantum chemical PM6 method augmented by dispersion and H-bonding correction terms reliably describes various types of noncovalent complexes. J. Chem. Theory Comput. 5, 1749–1760 (2009)
Stewart, J.J.P.: MOPAC 2012 Stewart Computational Chemistry: Colorado Springs, CO, USA
Rezac, J.: Cuby: an integrative framework for computational chemistry. J. Comput. Chem. 37, 1230–1237 (2016)
Becke, A.D.: New mixing of Hartree-Fock and local density-functional theories. J. Chem. Phys. 98, 1372–1377 (1993)
Chai, J.D., Head-Gordon, M.: Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem., Chem. Phys. 10, 6615–6620 (2008)
Zhao, Y., Truhlar, D.G.: The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 120, 215–241 (2008)
Møller, C., Plesset, M.S.: A note on an approximation treatment for many-electron systems. Phys. Rev. 46, 618–622 (1934)
Schlegel, H.B.: Potential energy curves using unrestricted Moller-Plesset perturbation theory with spin annihilation. J. Chem. Phys. 84, 4530 (1986)
Mayer, I.: Spin-projected UHF method. IV. Comparison of potential curves given by different one-electron methods. Adv. Quantum Chem. 12, 189–262 (1980)
Knowles, P.J., Andrews, J.S., Amos, R.D., Handy, N.C., Pople, J.A.: Restricted Møller-Plesset theory for open shell molecules. Chem. Phys. Lett. 186, 130–136 (1991)
Chan, B., Radom, L.: Assessment of theoretical procedures for hydrogen atom abstraction by chlorine, and related reactions. Theor. Chem. Acc. 130, 251–260 (2011)
Furche, F., Ahlrichs, A.: Adiabatic time-dependent density functional methods for excited state properties. J. Chem. Phys. 117, 7433–7447 (2002)
Turecek, F.: Benchmarking electronic excitation energies and transitions in peptide radicals. J. Phys. Chem. A. 119, 10101–10111 (2015)
Barbatti, M., Ruckenbauer, M., Plasser, F., Pittner, J., Granucci, G., Persico, M., Lischka, H.: Newton-X: a surface-hopping program for nonadiabatic molecular dynamics. Wiley Interdisciplinary Reviews: Comput. Mol. Sci. 4, 26–33 (2014)
Barbatti, M., Aquino, A.J., Lischka, H.: The UV absorption of nucleobases: semiclassical ab initio spectra simulations. Phys. Chem., Chem. Phys. 12, 4959–4967 (2010)
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H.P., Izmaylov, A.F., Bloino, J., Zheng, G., Sonnenberg, J.L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery Jr., J.A., Peralta, J.E., Ogliaro, F., Bearpark, M., Heyd, J.J., Brothers, E., Kudin, K.N., Staroverov, V.N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Rega, N., Millam, J.M., Klene, M., Knox, J.E., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Martin, R.L., Morokuma, K., Zakrzewski, V.G., Voth, G.A., Salvador, P., Dannenberg, J.J., Dapprich, S., Daniels, A.D., Farkas, O., Foresman, J.B., Ortiz, J.V., Cioslowski, J., Fox, D.J.: Gaussian 09, Revision A.02. Gaussian, Inc, Wallingford (2009)
Gilbert, R.G., Smith, S.C.: Theory of unimolecular and recombination reactions, pp. 52–132. Blackwell Scientific Publications, Oxford (1990)
Zhu, L., Hase, W.L.: Quantum chemistry program exchange; Indiana University: Bloomington (1994) Program no. QCPE 644
Frank, A.J., Sadílek, M., Ferrier, J.G., Turecek, F.: Sulfur oxyacids and radicals in the gas phase. A variable-time neutralization-photoexcitation-reionization mass spectrometric and ab initio/RRKM study. J. Am. Chem. Soc. 119, 12343–12353 (1997)
Pepin, R., Tureček, F.: Kinetic ion thermometers for electron transfer dissociation. J. Phys. Chem. B. 119, 2818–2826 (2015)
Tureček, F.: Proton affinity of dimethyl sulfoxide and relative stabilities of C2H6OS molecules and C2H7OS+ ions. A comparative G2(MP2) ab initio and density functional theory study. J. Phys. Chem. A. 102, 4703–4713 (1998)
Rablen, P.R.: Is the acetate anion stabilized by resonance or electrostatics? A systematic structural comparison. J. Am. Chem. Soc. 122, 357–368 (2000)
Tureček, F.: Temperature effects in mass spectrometry. On the estimation of thermal energies of organic molecules. Org. Mass Spectrom. 26, 1074–1081 (1991)
Turecek, F.: N–Cα-bond dissociation energies and kinetics in amide and peptide radicals. Is the dissociation a nonergodic process? J. Am. Chem. Soc. 125, 5954–5963 (2003)
Tureček, F., Syrstad, E.A., Seymour, J.L., Chen, X., Yao, C.: Peptide cation-radicals. A computational study of the competition between peptide N–Cα bond cleavage and loss of the side chain in the [GlyPhe-NH2 + 2H]+• cation radical. J. Mass Spectrom. 38, 1093–1104 (2003)
Pepin, R., Layton, E.D., Liu, Y., Afonso, C., Tureček, F.: Where does the electron go? Stable and metastable peptide cation radicals formed by electron transfer. J. Am. Soc. Mass Spectrom. 28, 164–181 (2017)
Acknowledgments
Support by the National Science Foundation Division of Chemistry (grants CHE-1359810, CHE-1661815, and CHE-1624430), and Klaus and Mary Ann Saegebarth Endowment are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 1606 kb)
Rights and permissions
About this article
Cite this article
Imaoka, N., Houferak, C., Murphy, M.P. et al. Spontaneous Isomerization of Peptide Cation Radicals Following Electron Transfer Dissociation Revealed by UV-Vis Photodissociation Action Spectroscopy. J. Am. Soc. Mass Spectrom. 29, 1768–1780 (2018). https://doi.org/10.1007/s13361-017-1871-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1871-0