Abstract
Vitamin D compounds are secosteroids, which are best known for their role in bone health. More recent studies have shown that vitamin D metabolites and catabolites such as dihydroxylated species (e.g., 1,25- and 24,25-dihydroxyvitamin D3) play key roles in the pathologies of various diseases. Identification of these isomers by mass spectrometry is challenging and currently relies on liquid chromatography, as the isomers exhibit virtually identical product ion spectra under collision induced dissociation conditions. Here, we developed a simple MALDI-CID method that utilizes ion activation of reactive analyte/matrix adducts to distinguish isomeric dihydroxyvitamin D3 species, without the need for chromatography separation or chemical derivatization techniques. Specifically, reactive 1,5-diaminonaphthalene adducts of dihydroxyvitamin D3 compounds formed during MADI were activated and specific cleavages in the secosteroid’s backbone structure were achieved that produced isomer-diagnostic fragment ions.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term “vitamin D” refers to a group of fat-soluble secosteroids, the most important members of which are vitamin D3, occurring in mammals, and D2 in plants (in particular mushrooms). The chemical structure of vitamin D was first elucidated by Adolf Windaus in 1935 [1], while the vital role of vitamin D in bone mineralization and calcium/phosphate absorption has been known for almost 100 years [2]. More recently, vitamin D deficiency has been linked to a much wider range of diseases, including diabetes, cancer, depression, neurodegenerative, and cardiovascular diseases [3,4,5]. It has been shown that the vitamin D receptor is present in most human organs and cells [6, 7], which has led to a surge of vitamin D-related research in the last two decades [8].
Vitamin D synthesis in humans starts with sunlight photosynthesis in the skin by conversion of 7-dehydrocholesterol (pro-vitamin D3) to previtamin D3, which is then transformed to vitamin D3 by thermal isomerization [9]. Photosynthesis requires ultraviolet radiation of wavelengths between 290 and 315 nm [6]. Because of the risk of skin cancers, limited sunlight exposure has led to an estimated one billion vitamin D deficient or insufficient people worldwide [6]. Metabolism of vitamin D3 in humans is extensive and over 50 metabolites have been reported in the literature [10]. After photosynthesis, vitamin D3 enters the circulation and is transported to the liver via vitamin D binding protein. In the liver, it undergoes oxidation to 25-hydroxyvitamin-D3 (25(OH)D3). Although 25(OH)D3 is the most abundant circulating metabolite of vitamin D3, it is biologically inactive. 25(OH)D3 is further hydroxylated in the kidney to produce the active form, 1,25-dihydroxyvitamin D3 [1,25(OH)2D]). This species is present at very low levels (picomolar range) in human blood, however, and exhibits very short blood half-life of only ~4 h, thus limiting the analytical utility as status marker [11]. Renal production of 1,25(OH)2D3 is regulated through feedback mechanisms and total amounts of metabolites are controlled by enzymatic degradations. That is, 25(OH)D3 and 1,25(OH)2D3 are further hydroxylated to 24,25(OH)2D3 and 1,24,25(OH)3D3 for excretion [12]. Interestingly, 24,25(OH)2D3 has been suggested as active metabolite with biological effects on its own [13,14,15].
Most vitamin D measurements today are performed by immunoassay techniques [16]. Recently, liquid chromatography-tandem mass spectrometry (LC-MS/MS) has emerged as the gold standard technique for vitamin D analysis because of its superior sensitivity and specificity [8, 17, 18]. However, due to the lipophilic nature of vitamin D compounds, abundant isobaric/isomeric interferences in biological fluids [19], and the low concentrations for many downstream vitamin D metabolites, most MS assays are restricted to the most abundant circulating 25(OH)D3 form [18]. This limitation can be overcome by chemical derivatization using reagents such as 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD) [20, 21] or Amplifex [15], which not only increase ionization efficiency but also decrease isobaric interference levels by shifting the m/z range of vitamin D compounds to higher values [22]. However, additional sample preparation steps are required, which are laborious and may reduce recovery values [22].
In this study, we were concerned with dihydroxylated isomers of vitamin D3, as they exhibit identical m/z for their protonated molecules, e.g., after electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI). They also generate virtually identical product ion spectra under collision-induced dissociation (CID) conditions, showing primarily [M+H−H2O]+ and [M+H−2H2O]+ product ions at low collision energies (Figure 1). At higher energies, numerous product ions are formed after the initial dehydrations, giving typical “picket fence” series of hydrocarbon chain decompositions, which do not provide structure-specific ions ([23] and Figure 9 in Reference [8]). Protocols that distinguish dihydroxylated vitamin D metabolites therefore require LC separation, often combined with chemical derivatization [15]. In addition, Yost and coworkers have recently successfully demonstrated the application of ion mobility spectrometry to distinguish epimeric species of 25(OH)D3 prior to mass spectrometric analysis, by utilizing sodiated species [24, 25].
To further explore direct mass spectrometry for distinguishing dihydroxylated vitamin D isomers – without the need for chromatography separation – a new MALDI method based on reactive analyte/matrix adducts was investigated here. Reactive analyte/matrix adducts were previously observed during matrix-assisted laser desorption/ionization (MALDI), where fragmentation occurred following laser-induced chemical reactions in the hot plume. These in-source decay (ISD) and post-source decay (PSD) techniques were successfully applied to protein sequencing because of their high sensitivities and tolerance of inhomogeneities [26]. Moreover, hydrogen-donor matrices such as 1,5-diaminonaphthalene (1,5-DAN) are able to directly catalyze hydrogen transfers between matrix and analyte prior to desorption [27,28,29,30], and thus induces N–Cα bond cleavages of the protein backbone, leading to c and z fragment ions similar to electron capture dissociation (ECD) [31]. In this study, we applied MALDI in combination with a hydrogen donor matrix to dihydroxyvitamin D3 isomers, namely 1,25(OH)2D3 and 24,25(OH)2D3. Ion activation of the reactive analyte/matrix adducts generated more structure informative product ions compared with conventional ESI-CID experiment, enabling the two isomers to be readily differentiated without any prior chemical derivatization or chromatographic separation.
Experimental
Reagents and Chemicals
1,25-Dihydroxyvitamin D3, 24,25-dihydroxyvitamin D3, 1,5-diaminonaphthalene (DAN), acetonitrile (ACN), and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (Steinheim, Germany). Vitamin D-free human serum was purchased from Golden West Biologicals (Temecula, CA, USA). The 96-well AC micro-extraction plates were from Tecan (Männedorf, Switzerland). Purified water was generated by a Millipore (Bedford, MA, USA) Direct-Q8 system. The standards were dissolved in H2O/ACN (50:50, v/v) at 20 μM concentration (stock solutions); 1,5-DAN (>20 mg/mL) was dissolved in H2O/ACN/TFA (50:50:0.1, v/v/v) to give a saturated solution.
Serum Preparation
Antibody-purified, vitamin D-free serum was fortified with the investigated dihydroxylated vitamin D species, viz. 1,25(OH)2D3 and 24,25(OH)2D3. The fortified samples were then extracted by supported liquid extraction (SLE) using 96-well micro-extraction plates, as previously described [32, 33].
Mass Spectrometry
Each analyte solution was mixed with 1,5-DAN solution at a ratio of 1:1 (v/v), deposited onto the steel MALDI target plates (Bruker, Bremen, Germany), and allowed to dry at ambient conditions. Mass spectra were recorded using a Bruker 7 Tesla Fourier transform ion cyclotron resonance MS, equipped with a frequency-tripled Nd:YAG laser. The laser power was optimized to obtain spectra with high signal-to-noise ratios (S/N). For spectra acquired from analytical standards, a single transient was recorded from 200 summed laser shots. For serum extracts, 40 transients were co-added to enhance S/N [34]. In MS/MS mode, precursor ions were isolated first in the quadrupole, externally accumulated in the hexapole for 0.1s, and 5–20 eV collision energy was applied for CID.
Results and Discussion
As mentioned in the Introduction, collision induced dissociation (CID) analyses of [M+H]+ ions of the dihydroxylated species 1,25(OH)2D3 and 24,25(OH)2D3 by electrospray ionization (ESI) gave virtually identical product ion spectra, for both low or high collision energies (Figure 1), prompting us to investigate alternative means of ion activation to generate structure-specific ions. Specifically, we investigated the utility of using reactive analyte/matrix adduct ions, promoted by MALDI hydrogen donor matrices, in combination with CID. Hydrogen donor matrices have previously been shown to induce c and z fragment ions with peptides, which was different from the conventional CID outcome during protein sequencing [26]. Among the various matrices, 1,5-diaminonaphthalene (1,5-DAN) was chosen in our study, as it has previously demonstrated strong hydrogen-donating abilities and is currently regarded to be the most efficient matrix for subsequent in-source decay (ISD) analyses [29]. Recently, Traldi and co-workers have investigated the ionization behavior of 1,5-DAN in detail and have shown that this matrix primarily generates radical [1,5-DAN]+• cations under MALDI conditions with unique abilities during analysis of proteins, i.e., possibility to activate ISD as well as reduction reactions of disulphide bonds [35].
Method Development
In the first set of experiments, MALDI full scan analyses revealed the expected [M+H]+ precursor species for 1,25(OH)2D3 and 24,25(OH)2D3 at m/z 417, similar to the ESI measurements. Not surprisingly, CID of these ions yielded the same product ion spectra as seen in ESI-CID experiments, exhibiting no isomer-specific ions. In addition to the protonated precursors, however, we also observed 1,5-DAN adduct ions, specifically [M+1,5-DAN]+• radical ions at m/z 574 for both investigated vitamin D3 metabolites. Unfortunately, we did not obtain any fragment ions from ISD or PSD processes from these reactive analyte/matrix adducts. In addition to the [M+1,5-DAN]+• species, protonated species as well product ions after single and double hydrogen abstraction were also seen in the spectra, demonstrating the unique reactivity of the 1,5-DAN matrix. For a detailed inspection, the [M+1,5-DAN]+• species was re-analyzed using FTICR-MS narrowband mode centred on m/z 575.0 with a 10 u window for enhanced mass resolution (Figure S1, Supplementary Material). A closer inspection of the [M+1,5-DAN+H]+ species revealed a composite of three ions, viz., C35H53N2O3 13C2 from [M+1,5-DAN-H]+, C36H54N2O3 13C1 from [M+1,5-DAN]+•, and C37H55N2O3 from [M+1,5-DAN+H]+ (Figure S1 inset, Supplementary Material).
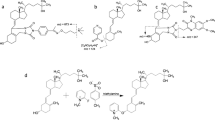
In the next experiment, we isolated the main [M+1,5-DAN]+• precursor ions after MALDI and performed CID to activate the complex. Dissociation of the [M+1,5-DAN]+• adduct was dominated by the loss of a neutral [1,5-DAN–H]• radical, primarily creating the [M+H]+ product ion, along with a minor ion signal for the [M]+• ion of 1,25(OH)2D3 and 24,25(OH)2D3 (Figure 2). That is, for the [M+H]+ ion, the described two-stage sequence of analyte/1,5-DAN adduct formation via MALDI, followed by isolation, activation, and dissociation of this reactive adduct by CID corresponded to the overall reaction
which is effectively the same net reaction as previously described for the proton-donating abilities of 1,5-DAN and [M+H]+ formation during MALDI of proteins [35].
CID mass spectra of [M+1,5-DAN]+• species of 1,25(OH)2D3 (top) and 24,25(OH)2D3 (bottom) (concentration, 10 μM ea.). The diagnostic product ions are highlighted in grey, which are further expanded in Figures 3 and 4, S2, S3. Inset: expansion of the 1,5-DAN isotopic peak pattern, showing a combination C9H10N2 13C1 and C10H11N2 species
Except for minor intensity differences, the distribution of product ions appeared at first very similar for 1,25(OH)2D3 and 24,25(OH)2D3, i.e., protonated dihydroxyvitamin D3 was formed along with a series of H2O losses. Closer inspection of the spectra revealed two unique fragment ions, however, which are highlighted Figure 2, namely, m/z 297 and 433 for 1,25(OH)2D3 (a mass scale expansion of these species is illustrated in Figure 3), and m/z 281 and 449 for 24,25(OH)2D3 (Figures S2 and S3, Supplementary Material). These ions were then examined in more detail by means of high resolution FTICR-MS with sub-ppm level mass accuracy. Figure 3a illustrates the expanded peak area for m/z 297 and the measured accurate mass unambiguously identified this signal as C18H21N2O2. This fragment ion can be rationalized via backbone cleavage of the single bond at C-6/C-7 of 1,25(OH)2D3, leaving the fragment ion with the 1,5-DAN adduct attached to the C-6 side of the molecule. This peak was specific to 1,25(OH)2D3, as the observed fragment ion contained two hydroxyl groups in ring A, whereas the corresponding product ions for 24,25(OH)2D3 only possesses a single –OH group in the A-ring moiety of the secosteroid. Consequently, no signal was seen for 24,25(OH)2D3 at the relevant m/z range (Figure 3a, middle spectrum). The second product ion at m/z 433 was identified as C29H41N2O, resulting from the same C-6/C-7 cleavage, with 1,5-DAN and the charge attached to the C-7 side of the molecule (Figure 3b), which provided complementary proof of the backbone bond dissociation of C-6/C-7.
Mass scale expansion of Figure 2: (a) m/z ~297–299, and (b) m/z ~433–435 for CID fragment ions of 1,25(OH)2D3 (top traces) and 24,25(OH)2D3 (middle traces) (concentration, 10 μM ea.). The bottom spectra show the isotope simulations of the assigned elemental formulae
In LC-MS/MS assays, the simple H2O losses during in-source CID or after subsequent ion activation make unambiguous structural identification of hydroxylated vitamin D3 isomers based solely on mass spectrometry virtually impossible [8]. We have previously shown that the diagnostic value of product ions resulting from dehydration reaction is very limited for vitamin D compounds because of multiple isobaric interferences present in biofluids, several of which exhibit identical water losses upon CID [23]. In addition, using higher collision energies does not create new diagnostic backbone ions, as the initial dehydrations are followed by multiple parallel reaction series from direct C-C cleavages, rearrangements, and unsaturations [23], thus generating very similar product ion spectra for all vitamin D metabolites. Owing to these specificity limitations, regular vitamin D metabolite assays require chromatographic retention time as additional identification criterion [8].
The particular MS/MS behavior observed here after 1,5-DAN addition was different, however, as the fragile OH groups were preserved during dissociation of the C-6/C-7 bond, making the determination of individual vitamin D isomers readily possible. It was equally interesting to observe that 1,5-DAN remained attached to both possible fragment ions during CID, as illustrated in Figure 3, with no preference of 1,5-DAN binding to a certain location of the dihydroxylated vitamin D3 isomers. To shed further light on this process, we examined the 1,5-DAN-related product ions in more detail. The 1,5-DAN-related product ions were visible at m/z 158 ([1,5-DAN]+•) and a smaller signal at m/z 159. The latter peak was comprised of two species separated by 4.8 mu, which the FTICR-MS instrument was readily able to resolve: (1) the 13C contribution from [1,5-DAN]+• (C9H10N2 13C1) and (2) protonated 1,5-DAN (C10H11N2). The hydrogen transfer between the fragment ions suggests that hydrogen bonding exists between the two compounds. Considering the heterocyclic nitrogen of 1,5-DAN and the hydroxyl groups of 1,25(OH)2D3, formation of O–H···N and N–H···O bonds was considered. We suggest that the hydrogen bonding occurs during the MALDI process [29], stabilizing and preventing the analyte/matrix adduct from dissociation during MS/MS. The same experiment was subsequently repeated for 24,25(OH)2D3, and the same result was seen for the C-6/C-7 dissociation (Figures S2 and S3, Supplementary Material).
In the subsequent experiment, 1,25(OH)2D3 and 24,25(OH)2D3 standards were mixed at 1:1 ratio for MALDI-CID measurements, to directly compare the relative abundances of the two dihydroxyvitamin D3 species. Characteristic product ions were chosen, i.e., m/z 297 for 1,25(OH)2D3 (Figure 3a) and m/z 281 for 24,25(OH)2D3 (Figure S2, Supplementary Material), and peak areas of the two selected ions were recorded and their ratios calculated. As seen in Table S2 (Supplementary Material), the fragment ratios from the 1:1 mixtures of 1,25(OH)2D3 and 24,25(OH)2D3 were measured with an average ratio of 2.9 (6.9% RSD), indicating the general ability for relative quantification in a single CID experiment. The reproducibility of the measured ion currents (intra-spot) was very good (≤10% RSD), as no laser rastering was performed during acquisition to avoid sweet spot phenomena. Using dedicated isotopologues of the vitamin D3 metabolites as internal standards, which will exhibit identical adduct formation, would provide the possibility for absolute quantification.
Measurement of Serum Sample
The proof-of-concept work presented in the previous section was utilized to attempt separation of dihydroxylated vitamin D metabolites in human serum samples. The product ions of the MALDI adducts provided unique specificity for isomer separation, as shown above; however, the dihydroxylated vitamin D3 metabolites exist at very low concentrations in human serum, in particular 1,25(OH)2D3, with concentration levels in the picomolar range [32]; 24,25(OH)2D3 levels are higher, typically one order of magnitude below those of 25(OH)D3, that is, in the low ng/mL range [32]. Since both species are simultaneously present in serum, sufficient detection sensitivity was required here to capture both the lower abundant 1,25(OH)2D3 species as well as the 24,25(OH)2D3 compound.
For these experiments, we utilized antibody-purified, vitamin D-free human serum and spiked the samples with 1,25(OH)2D3 (at 20 ng/mL) and 24,25(OH)2D3 (at 30 ng/mL) and measured the sample using the same experimental parameters described above. As seen in Figure 4, various concomitant isobaric interferences from the serum matrix were present, but the characteristic ions for 1,25(OH)2D3 and 24,25(OH)2D3 are observed at m/z ~297 and m/z ~281 with relative peak areas of 0.32% and 0.19%, respectively. Considering the fragment ion ratio measured above, the relative abundance of 1,25(OH)2D3 and 24,25(OH)2D3 was determined as ~0.63. This slight deviation from the fortified ratio could likely be improved by using internal standards.
MALDI-CID measurement of human serum sample using the same experimental parameters as shown in Figure 2 (concentrations, 20 ng/mL for 1,25(OH)2D3; 30 ng/mL for 24,25(OH)2D3). The characteristic product ions of 1,25(OH)2D3 (m/z: 297.1597, ▲) and 24,25(OH)2D3 (m/z: 281.1647, ●) were expanded in the insets
It is evident that the detection sensitivity for 1,25(OH)2D3 was not sufficient using the described method, with observed limits of quantification approximately 100-fold higher than required for physiological levels. It is important to consider, however, that we utilized serum volumes of only 50 μL, combined with an established high throughput sample extraction protocol for the vitamin D metabolites, as previously described for LC-MS/MS [32, 33]. That is, the method could be improved if a dedicated sample preparation routine is developed, implementing higher analyte enrichment factors and/or larger samples volumes. Alternatively, a more sensitive mass spectrometry platform could be chosen for quantitative experiments. Ideally, a MALDI-triple quadrupole (QqQ) platform is used, utilizing the dedicated high duty cycle multiple reaction monitoring (MRM) mode, which would provide high sensitivity and high precision analyses. We have previously shown that a dedicated MALDI-QqQ instrument using specific MRM transitions was able to determine pharmaceutical drugs at limits of quantification in the picomolar range [36].
Conclusions
In the present study, a fast and simple MALDI method was developed to identify dihydroxylated vitamin D3 isomers. CID experiments of dihydroxyvitamin D3 compounds using reactive 1,5-DAN adducts provided specific cleavages at C-6 and C-7 of the secosteroid’s backbone structures and produced structure-diagnostic fragment ions, which enabled unambiguous differentiation of 1,25(OH)2D3 and 24,25(OH)2D3. The same protocol would be equally applicable to other hydroxylated compounds such as oxysterols. The reported method was demonstrated for use with MALDI and could therefore be readily extended to mass spectrometry imaging applications, for mapping the distribution of various isomeric vitamin D metabolites across tissue surfaces. Moreover, in principle, it should be possible to adapt the reactive MALDI method to other direct mass spectrometry techniques such as ambient ionization or direct infusion/flow injection analyses, by using appropriate chemical modifiers to condition the precursor molecules followed by MS/MS.
References
Windaus, A., Thiele, W.: Über die Konstitution des Vitamins D2. Justus Liebigs Ann. Chem. 521, 160–175 (1936)
Wolf, G.: The discovery of vitamin D: the contribution of Adolf Windaus. J. Nutr. 134, 1299–1302 (2004)
Holick, M.F.: Vitamin D and sunlight: strategies for cancer prevention and other health benefits. Clin. J. Am. Soc. Nephrol. 3, 1548–1554 (2008)
Shipowick, C.D., Moore, C.B., Corbett, C., Bindler, R.: Vitamin D and depressive symptoms in women during the winter: a pilot study. Appl. Nurs. Res. 22, 221–225 (2009)
Gueli, N., Verrusio, W., Linguanti, A., Di Maio, F., Martinez, A., Marigliano, B., Cacciafesta, M.: Vitamin D: drug of the future. A new therapeutic approach. Arch. Gerontol. Geriatr. 54, 222–227 (2012)
Holick, M.F.: Vitamin D seficiency. N. Engl. J. Med. 357, 266–281 (2007)
Stokes, C.S., Volmer, D.A., Grünhage, F., Lammert, F.: Vitamin D in chronic liver disease. Liver Int. 33, 338–352 (2013)
Volmer, D.A., Mendes, L.R.B.C., Stokes, C.S.: Analysis of vitamin D metabolic markers by mass spectrometry: current techniques, limitations of the “gold standard” method, and anticipated future directions. Mass Spectrom. Rev. 34, 2–23 (2015)
Holick, M.F., MacLaughlin, J.A., Clark, M.B., Holick, S.A., Potts, J.T., Anderson, R.R., Blank, I.H., Parrish, J.A., Elias, P.: Photosynthesis of previtamin D3 in human skin and the physiologic consequences. Science. 210, 203-205 (1980)
Zerwekh, J.E.: Blood biomarkers of vitamin D status. Am. J. Clin. Nutr. 87, 1087S–1091S (2008)
Crawford, B.A., Labio, E.D., Strasser, S.I., McCaughan, G.W.: Vitamin D replacement for cirrhosis-related bone disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 689–699 (2006)
Beckman, M.J., Tadikonda, P., Werner, E., Prahl, J., Yamada, S., DeLuca, H.F.: Human 25-hydroxyvitamin D3-24-hydroxylase, a multicatalytic enzyme. Biochemistry. 35, 8465–8472 (1996)
Endo, H., Kiyoki, M., Kawashima, K., Naruchi, T., Hashimoto, Y.: Vitamin D3 metabolites and PTH synergistically stimulate bone formation of chick embryonic femur in vitro. Nature. 286, 262–264 (1980)
Seo, E.-G., Norman, A.W.: Three-fold induction of renal 25-hydroxyvitamin D3-24-hydroxylase activity and increased serum 24,25-dihydroxyvitamin D3 levels are correlated with the healing process after chick tibial fracture. J. Bone Miner. Res. 12, 598–606 (1997)
Müller, M.J., Stokes, C.S., Volmer, D.A.: Quantification of the 3α and 3β epimers of 25-hydroxyvitamin D3 in dried blood spots by LC-MS/MS using artificial whole blood calibration and chemical derivatization. Talanta. 165, 398–404 (2017)
Heijboer, A.C., Blankenstein, M.A., Kema, I.P., Buijs, M.M.: Accuracy of six routine 25-hydroxyvitamin D assays: influence of vitamin D binding protein concentration. Clin. Chem. 58, 543-548 (2012)
El-Khoury, J.M., Reineks, E.Z., Wang, S.: Progress of liquid chromatography-mass spectrometry in measurement of vitamin D metabolites and analogues. Clin. Biochem. 44, 66–76 (2011)
Müller, M.J., Volmer, D.A.: Mass spectrometric profiling of vitamin D metabolites beyond 25-hydroxyvitamin D. Clin. Chem. 61, 1033-1048 (2015)
Qi, Y., Geib, T., Schorr, P., Meier, F., Volmer, D.A.: On the isobaric space of 25-hydroxyvitamin D in human serum: potential for interferences in liquid chromatography/tandem mass spectrometry, systematic errors and accuracy issues. Rapid Commun. Mass Spectrom. 29, 1–9 (2015)
Eyles, D., Anderson, C., Ko, P., Jones, A., Thomas, A., Burne, T., Mortensen, P.B., Nørgaard-Pedersen, B., Hougaard, D.M., McGrath, J.: A sensitive LC/MS/MS assay of 25OH vitamin D3 and 25OH vitamin D2 in dried blood spots. Clin. Chim. Acta. 403, 145–151 (2009)
Higashi, T., Shibayama, Y., Fuji, M., Shimada, K.: Liquid chromatography-tandem mass spectrometric method for the determination of salivary 25-hydroxyvitamin D3: a noninvasive tool for the assessment of vitamin D status. Anal. Bioanal. Chem. 391, 229–238 (2008)
Ding, S., Schoenmakers, I., Jones, K., Koulman, A., Prentice, A., Volmer, D.A.: Quantitative determination of vitamin D metabolites in plasma using UHPLC-MS/MS. Anal. Bioanal. Chem. 398, 779–789 (2010)
Duan, X., Weinstock-Guttman, B., Wang, H., Bang, E., Li, J., Ramanathan, M., Qu, J.: Ultrasensitive quantification of serum vitamin D metabolites using selective solid-phase extraction coupled to microflow liquid chromatography and isotope-dilution mass spectrometry. Anal. Chem. 82, 2488–2497 (2010)
Chouinard, C.D., Cruzeiro, V.W.D., Beekman, C.R., Roitberg, A.E., Yost, R.A.: Investigating differences in gas-phase conformations of 25-hydroxyvitamin D3 sodiated epimers using ion mobility-mass spectrometry and theoretical modeling. J. Am. Soc. Mass Spectrom. 28, 1497–1505 (2017)
Chouinard, C.D., Cruzeiro, V.W.D., Roitberg, A.E., Yost, R.A.: Experimental and theoretical investigation of sodiated multimers of steroid epimers with ion mobility-mass spectrometry. J. Am. Soc. Mass Spectrom. 28, 323–331 (2017)
Hardouin, J.: Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry. Mass Spectrom. Rev. 26, 672–682 (2007)
Köcher, T., Engström, Å., Zubarev, R.A.: Fragmentation of peptides in MALDI in-source decay mediated by hydrogen radicals. Anal. Chem. 77, 172–177 (2005)
Asakawa, D., Calligaris, D., Smargiasso, N., De Pauw, E.: Ultraviolet laser induced hydrogen transfer reaction: study of the first step of MALDI in-source decay mass spectrometry. J. Phys. Chem. B. 117, 2321–2327 (2013)
Demeure, K., Quinton, L., Gabelica, V., De Pauw, E.: Rational selection of the optimum MALDI matrix for top-down proteomics by in-source decay. Anal. Chem. 79, 8678–8685 (2007)
Asakawa, D., Smargiasso, N., De Pauw, E.: Estimation of peptide N–Cα bond cleavage efficiency during MALDI-ISD using a cyclic peptide. J. Mass Spectrom. 51, 323–327 (2016)
Qi, Y., Volmer, D.A.: Electron-based fragmentation methods in mass spectrometry: an overview. Mass Spectrom. Rev. 36, 4–15 (2017)
Müller, M.J., Stokes, C.S., Lammert, F., Volmer, D.A.: Chemotyping the distribution of vitamin D metabolites in human serum. Sci Rep. 6, 21080 (2016)
Geib, T., Meier, F., Schorr, P., Lammert, F., Stokes, C.S., Volmer, D.A.: A simple micro-extraction plate assay for automated LC-MS/MS analysis of human serum 25-hydroxyvitamin D levels. J. Mass Spectrom. 50, 275–279 (2015)
Qi, Y., O’Connor, P.B.: Data processing in Fourier transform ion cyclotron resonance mass spectrometry. Mass Spectrom. Rev. 33, 333–352 (2014)
Molin, L., Seraglia, R., Dani, F.R., Moneti, G., Traldi, P.: The double nature of 1,5-diaminonaphthalene as matrix-assisted laser desorption/ionization matrix: some experimental evidence of the protonation and reduction mechanisms. Rapid Commun. Mass Spectrom. 25, 3091–3096 (2011)
Sleno, L., Volmer, D.A.: Some fundamental and technical aspects of the quantitative analysis of pharmaceutical drugs by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 19, 1928–1936 (2005)
Acknowledgments
This work was supported by the German Research Foundation (DFG VO 1355/4-1 and FTICR-MS Facility, INST 256/356-1). D.A.V. acknowledges general research support by the Alfried Krupp von Bohlen und Halbach-Stiftung.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 948 kb)
Rights and permissions
About this article
Cite this article
Qi, Y., Müller, M.J. & Volmer, D.A. Activation of Reactive MALDI Adduct Ions Enables Differentiation of Dihydroxylated Vitamin D Isomers. J. Am. Soc. Mass Spectrom. 28, 2532–2537 (2017). https://doi.org/10.1007/s13361-017-1775-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1775-z