Abstract
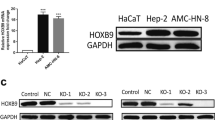
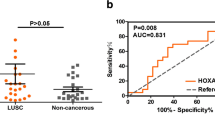
Laryngeal squamous cell carcinoma (LSCC) is a very aggressive cancer, considered to be a subtype of the head and neck squamous cell carcinoma (HNSCC). Despite significant advances in the understanding and treatment of cancer, prognosis of patients with LSCC has not improved recently. In the present study, we sought to understand better the genetic mechanisms underlying LSCC development. Thirty-two tumor samples were collected from patients undergoing surgical resection of LSCC. The samples were submitted to whole-genome cDNA microarray analysis aiming to identify genetic targets in LSCC. We also employed bioinformatic approaches to expand our findings using the TCGA database and further performed functional assays, using human HNSCC cell lines, to evaluate viability, cell proliferation, and cell migration after silencing of selected genes. Eight members of the homeobox gene family (HOX) were identified to be overexpressed in LSCC samples when compared to normal larynx tissue. Quantitative RT-PCR analysis validated the overexpression of HOX gene family members in LSCC. Receiver operating characteristic (ROC) statistical method curve showed that the expression level of seven members of HOX gene family can distinguish tumor from nontumor tissue. Correlation analysis of clinical and gene expression data revealed that HOXC8 and HOXD11 genes were associated with the differentiation degree of tumors and regional lymph node metastases, respectively. Additionally, siRNA assays confirmed that HOXC8, HOXD10, and HOXD11 genes might be critical for cell colony proliferation and cell migration. According to our findings, several members of the HOX genes were overexpressed in LSCC samples and seem to be required in biological processes involved in tumor development. This suggests that HOX genes might play a critical role in the physiopathology of LSCC tumors.





Similar content being viewed by others
References
IARC – International Agency for Research on Cancer. Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. https://www.iarc.fr. Accessed in 21 Jul 2015.
Hardisson D. Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2003;260(9):502–8.
Zhang SY, ZM L, Luo XN, Chen LS, Ge PJ, Song XH, et al. Retrospective analysis of prognostic factors in 205 patients with laryngeal squamous cell carcinoma who underwent surgical treatment. PLoS One. 2013;8(4):e60157.
Silveira NJ, Varuzza L, Machado-Lima A, Lauretto MS, Pinheiro DG, Rodrigues RV, Severino P, Nobrega FG, Head and Neck Genome Project GENCAPO, Silva Jr WA, de B Pereira CA, Tajara EH. Searching for molecular markers in head and neck squamous cell carcinomas (HNSCC) by statistical and bioinformatic analysis of larynx-derived SAGE libraries. BMC Med Genet. 2008;11(1):56.
Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest. 2012;122(6):1951–7.
Colombo J, Fachel AA, De Freitas Calmon M, Cury PM, Fukuyama EE, Tajara EH, Cordeiro JA, Verjovski-Almeida S, Reis EM, Rahal P. Gene expression profiling reveals molecular marker candidates of laryngeal squamous cell carcinoma. Oncol Rep. 2009;21(3):649–63.
Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris III HA, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–703.
Petrelli F, Borgonovo K, Cabiddu M, Lonati V, Barni S. Relationship between skin rash and outcome in non-small-cell lung cancer patients treated with anti-EGFR tyrosine kinase inhibitors: a literature-based meta-analysis of 24 trials. Lung Cancer. 2012;78(1):8–15.
Cai J, Ma H, Huang F, Zhu D, Bi J, Ke Y, Zhang T. Correlation of bevacizumab-induced hypertension and outcomes of metastatic colorectal cancer patients treated with bevacizumab: a systematic review and meta-analysis. World J Surg Oncol. 2013;28:11–306.
Gore L, DeGregori J, Porter CC. Targeting developmental pathways in children with cancer: what price success? Lancet Oncol. 2013;14(2):e70–8.
Plaça JR, Bueno RBL, Pinheiro DG, Panepucci RA, Araújo LF, Mamede RCM, Figueiredo DLA, Silva Jr WA. Gene expression analysis of laryngeal squamous cell carcinoma. Genomics Data. 2015;(5):9–12.
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot T, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;5(8):44–e71.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25(4):402–8.
Broad Institute TCGA Genome Data Analysis Center. Correlations between copy number and mRNAseq expression. Broad Institute of MIT and Harvard. 2016. doi:10.7908/C19886DH.
Broad Institute TCGA Genome Data Analysis Center. Correlation between mRNA expression and DNA methylation. Broad Institute of MIT and Harvard. 2016. doi:10.7908/C15M653H.
Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–71.
Miyamoto K, Fukutomi T, Akashi-Tanaka S, et al. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116:407–14.
Cai LY, Abe M, Izumi S, et al. Identification of PRTFDC1 silencing and aberrant promoter methylation of GPR150, ITGA8 and HOXD11 in ovarian cancers. Life Sci. 2007;80:1458–65.
Furuta J, Nobeyama Y, Umebayashi Y, et al. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6.
Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl). 2014;92:811–23.
Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Jin H, Si J. Homeobox D10 gene, a candidate tumor suppressor, is downregulated through promoter hypermethylation and associated with gastric carcinogenesis. Mol Med. 2012;9(18):389–400.
Vardhini NV, Rao PJ, Murthy PB, Sudhakar G. HOXD10 expression in human breast cancer. Tumor Biol. 2014;35(11):10855–60.
Sekar P, Bharti JN, Nigam JS, Sharma A, Soni PB. Evaluation of p53, HoxD10 and E-cadherin status in breast cancer and correlation with histological grade and other prognostic factors. J Oncol. 2014;2014:7025–7.
Osborne J, Hu C, Hawley C, Underwood LJ, O’Brien TJ, Baker VV. Expression of HOXD10 gene in normal endometrium and endometrial adenocarcinoma. J Soc Gynecol Investig. 1998;5(5):277–80.
Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie F, Lei L, Chen Y, Mao B, Jiang M, Li J, Wang D, Wang G. miR-22 promotion of cell migration and invasion by targeting homeobox D10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29(4):835–42.
Sharpe DJ, Orr KS, Moran M, White SJ, McQuaid S, Lappin TR, Thompson A, JA J. POUF2F1 activity regulates HOXD10 and HOXD11 promoting a proliferative and invasive phenotype in head and neck cancer. Oncotarget. 2014;5(18):8803–15.
Rodini CO, Xavier FC, Paiva KB, De Souza Setúbal Destro MF, Moyses RA, Michaluarte P, Carvalho MB, Fukuyama EE, Head and Neck Project Gencapo, Tajara EH, Okamoto OK, Nunes FD. Homeobox gene expression profile indicates HOXA5 as a candidate prognostic marker in oral squamous cell carcinoma. Int J Oncol. 2012;40(4):1180–8.
Adwan H, Zhivkova-Galunska M, Georges R, Eyol E, Kleeff J, Giese NA, Friess H, Bergmann F, Berger MR. Expression of HOXC8 is inversely related to the progression and metastasis of pancreatic ductal adenocarcinoma. Br J Cancer. 2011;105(2):288–95.
YB D, Dong B, Shen LY, Yan WP, Dai L, Xiong HC, Liang Z, Kang XZ, Qin B, Chen KN. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res. 2014;188(2):442–50.
Li Y, Chao F, Huang B, Liu D, Kim J, Huang S. HOXC8 promotes breast tumorigenesis by transcriptionally facilitating cadherin-11 expression. Oncotarget. 2014;5(9):2596–607.
Acknowledgments
We thank Amélia Goes de Araújo and Patrícia Vianna Bonini Palma for the technical support at Flow Cytometry Core at National Institute of Science and Technology in Stem Cell and Cell Therapy for the technical support. We also thank Josane de Freitas Sousa for the critical review of the manuscript final version. This work was supported by grant no. 140427/2010-4, National Counsel of Technological and Scientific Development (CNPq); grant no. 559809/2009-3 CNPq/GENOPROT; and grant nos. 2012/00588-5 and #2013/08135-2, São Paulo Research Foundation (FAPESP).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Ribeirão Preto Medical School, University of São Paulo (USP) (Proc. No. 9371/2003). All patients underwent surgical resection at the Head and Neck Surgery Division of the Department of Ophthalmology, Otorhinolaryngology and Head & Neck of Ribeirão Preto Medical School, USP, and an informed consent was obtained from patients before surgery.
Conflicts of interest
None.
Additional information
Rafaela de Barros e Lima Bueno and Anelisa Ramão contributed equally to this work.
Electronic supplementary material
ESM 1
(DOCX 32879 kb)
Rights and permissions
About this article
Cite this article
de Barros e Lima Bueno, R., Ramão, A., Pinheiro, D.G. et al. HOX genes: potential candidates for the progression of laryngeal squamous cell carcinoma. Tumor Biol. 37, 15087–15096 (2016). https://doi.org/10.1007/s13277-016-5356-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5356-8