Abstract
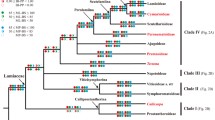
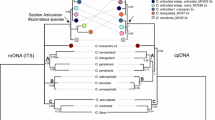
The phylogeny of the Acarosporaceae (Lecanoromycetes, Acarosporomycetidae, Acarosporales) is investigated using data from three molecular markers; nuclear ITS-LSU rDNA, mitochondrial SSU and β-tubulin. Acarosporaceae is shown to be constituted by six main clades; Myriospora, Timdalia, Pleopsidium, a clade composed by “Acarospora” rhizobola and “A.” terricola, the poorly supported Sarcogyne clade (including several Polysporina and Acarospora species) and the Acarospora clade (including the type of Polysporina, P. simplex, and several other Polysporina species). The common ancestor of the Acarosporaceae did not produce strongly black pigmented (carbonized or melanized) ascomata, but this trait has arisen secondarily and independently numerous times in the evolution of the group. The number of changes in character states of both carbonized epihymenium and carbonized exciple are considerably more than the minimum number. The genera Sarcogyne and Polysporina—largely circumscribed based on the presence of black pigmented ascomata—are shown to be distinctly non-monophyletic. The presence of green algae in the ascoma margin (lecanorine or lecideine ascomata) may vary even within single species.



Similar content being viewed by others
References
Ahti T, Brodo IM, Noble WJ (1987) Contributions to the lichen flora of British Columbia, Canada. Mycotaxon 28:91–97
Aptroot A (2002) New and interesting lichens and lichenicolous fungi in Brazil. Fungal Divers 9:15–45
Arcadia L, Knudsen K (2012) The name Myriospora is available for the Acarospora smaragdula group. Opuscula Philolich 11:19–25
Bendiksby M, Timdal E (2013) Molecular phylogenetics and taxonomy of Hypocenomyce sensu lato (Ascomycota: Lecanoromycetes): extreme polyphyly and morphological/ecological convergence. Taxon 62:940–956
Bollback JP (2006) SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinf 7:88
Brusse F (1988) Lithoglypha, a new lichen genus from Clarens Sandstone. Bothalia 18:89–96
Butler MJ, Day AW (1998) Fungal melanine: a review. Can J Microbiol 44:1115–1136
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Crewe AT, Purvis OW, Wedin M (2006) Molecular phylogeny of Acarosporaceae (Ascomycota) with focus on the proposed genus Polysporinopsis. Mycol Res 110:521–526
Ekman S, Andersen HL, Wedin M (2008) The limitation of ancestral state reconstruction and the evolution of the ascus in the Lecanorales (Lichenized Ascomycota). Syst Biol 57:141–156
Ekman S, Wedin M, Lindblom L, Jørgensen PM (2014) Extended phylogeny and a revised generic classification of the Pannariaceae (Peltigerales, Ascomycota). Lichenologist 46:627–656
Eriksson OE, Hawksworth DL (1991) Outline of the ascomycetes—1990. Syst Asc 9:39–271
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes, application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gauslaa Y, Solhaug KA (2001) Fungal melanins as a sun screen for symbiotic green algae in the lichen Lobaria pulmonaria. Oecologia 126:462–471
Goloboff P, Farris J, Nixon K (2008) TNT a free program for phylogenetic analysis. Cladistics 24:774–786
Gueidan C, Monnat J-Y, Navarro-Rosines P, Roux C (2014) Trimmatothelopsis versipellis—Découverte de stations dans le Finistére (France), position phylogénétique et consequences taxonomiques. Bull Soc linn Provence 65:47–65
Hafellner J (1993) Acarospora und Pleopsidium—zwei lichenisierte Ascomycetengattungen (Lecanorales) mit zahlreichen Konvergenzen. Nova Hedwigia 56:281–305
Hafellner J (1995) Towards a better circumscription of the Acarosporaceae (lichenized Ascomycotina Lecanorales). Cryptogam Bot 5:99–104
Hafellner J, Casares-Porcel M (1992) Untersuchungen an den Typusarten der lichenisierten Ascomycetengattungen Acarospora und Biatorella und die daraus entstehenden Konsequenzen. Nova Hedwigia 55:309–323
Hafellner J, Türk R (2001) Die lichenisierten Pilze Österreichs—eine Checkliste der bisher nachgewiesenen Arten mit verbreitungsangaben. Stapfia 76:1–167
Hauck M, Dulamsuren C, Mühlenberg M (2007) Lichen diversity on steppe slopes in the northern Mongolian mountain taiga and its dependence on microclimate. Flora 202:530–546
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Syst Biol 42:182–192
Huelsenbeck JP, Bollback JP (2001) Empirical and hierarchical Bayesian estimation of ancestral states. Syst Biol 50:351–366
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314
Huelsenbeck JP, Nielsen R, Bollback JP (2003) Stochastic mapping of morphological characters. Syst Biol 52:131–158
Jian S, Soltis PS, Gitzendanner MA, Moore MJ, Li R, Hendry TA, Qiu Y-L, Dhingra A, Bell CD, Soltis DE (2008) Resolving an ancient rapid radiation in Saxifragales. Syst Biol 57:38–57
Kantvilas G (1998) Notes on Polysporina Vězda with a description of a new species from Tasmania. Lichenologist 30:551–562
Kantvilas G, Seppelt RD (2006) Polysporina frigida sp. nov. from Antarctica. Lichenologist 38:109–113
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Katoh K, Toh H (2008a) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Katoh K, Toh H (2008b) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinf 9:212
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Knudsen K (2007a) Acarospora. In: Nash TH III, Gries C, Bungartz F (eds) Lichen Flora of the Greater Sonoran Desert Region, vol 3. Arizona State University, Tempe, Lichens Unlimited, pp 1–38
Knudsen K (2007b) P. In: Nash TH III, Gries C, Bungartz F (eds) Lichen Flora of the Greater Sonoran Desert Region, vol 3. Arizona State University, Tempe, Lichens Unlimited, pp 276–278
Knudsen K, Etayo J (2009) Sarcogyne algerica H. Magn., new to Europe. Opuscula Philolich 7:61–64
Knudsen K, Flakus A (2009) Acarospora ramosa (Acarosporaceae), a new effigurate yellow species from South America. Nova Hedwigia 89:349–353
Knudsen K, Fox HF (2010) Acarospora benedarensis: a rare terricolous maritime lichen from Ireland, Scotland, and Wales. Opuscula Philolich 9:31–34
Knudsen K, Kocourková J (2008) A study of lichenicolous species of Polysporina (Acarosporaceae). Mycotaxon 105:149–164
Knudsen K, Kocourková J (2010) Lichenological notes #1: Acarosporaceae. Mycotaxon 112:361–366
Knudsen K, Standley SM (2007) Sarcogyne. In: Nash TH III, Gries C, Bungartz F (eds) Lichen Flora of the Greater Sonoran Desert Region, vol 3. Arizona State University, Tempe, Lichens Unlimited, pp 289–296
Knudsen K, Lendemer JC, Harris RC (2010) Studies in lichens and lichenicolous fungi – no 15: miscellaneous notes on species from eastern North America. Opuscula Philolich 9:45–75
Lewis PO (2001) A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 50:913–925
Lewis PO, Holder MT, Holsinger KE (2005) Polytomies and Bayesian phylogenetic inference. Syst Biol 54:241–253
Lumbsch HT, Leavitt SD (2011) Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers 50:59–72
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org. Accessed 23 July 2014
Magnusson AH (1924) A monograph of the Scandinavian species of the genus Acarospora. Göteborgs Kungl Vetensk Samhälles Handl ser 4(28):1–150
Magnusson AH (1929) A monograph of the genus Acarospora. Kungl Svenska Vetenskapsakad Handl ser 3(7):1–400
Magnusson AH (1935) Acarosporaceae. Thelocarpaceae Rabenh Krypt-Fl 9 5:1–318
McLean J, Purvis OW, Williamson BJ, Bailey EH (1998) Role for lichen melanins in uranium remediation. Nature 391:649–650
Miadlikowska J, Kauff F, Högnabba F, Oliver JC, Molnár K, Fraker E, Gaya E, Hafellner J, Hofstetter V, Gueidan C, Kukwa M, Lücking R, Björk C, Sipman HJM, Burgaz AR, Thell A, Passo A, Myllys L, Goward T, Fernández-Brime S, Hestmark G, Lendemer J, Lumbsch HT, Schmull M, Schoch C, Sérusiaux E, Maddison DR, Arnold AE, Lutzoni F, Stenroos S (2014) A multigene phylogenetic synthesis for the class Lecanoromycetes (Ascomycota): 1307 fungi representing 1139 infrageneric taxa, 317 genera and 66 families. Mol Phylogenet Evol 79:132–168
Myllys L, Lohtander K, Tehler A (2001) β-tubulin, ITS and group I intron sequences challenge the species pair concept in Physcia aipolia and P. caesia. Mycologia 93:335–343
Navarro-Rosinés P, Roux C, Bellemère A (1999) Thelocarpella gordensis gen. et sp. nov. (Ascomycetes lichenisati, Acarosporaceae). Can J Bot 77:835–842
Nybakken L, Solhag KA, Bilger W, Gauslaa Y (2004) The lichens Xanthoria elegans and Cetraria islandica maintain a high protection against UV-B radiation in Arctic habitats. Oecologia 140:211–216
Otálora MAG, Aragón G, Martínez I, Wedin M (2013) Cardinal characters on a slippery slope – a re-evaluation of phylogeny character evolution and evolutionary rates in the jelly lichens (Collemataceae s.str.). Mol Phylogenet Evol 68:185–198
Otálora MAG, Jørgensen PM, Wedin M (2014) A revised generic classification of the jelly lichens, Collemataceae. Fungal Divers 64:275–293
Pagel M, Meade A (2006) Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov Chain Monte Carlo. Am Nat 167:808–825
Pagel M, Meade A (2007) BayesTraits Version 1.0 Computer Package. Software. http://www.evolution.reading.ac.uk/BayesTraits.html. Accessed 23 July 2014
Pagel M, Meade A, Barker D (2004) Bayesian estimation of ancestral states on phylogenies. Syst Biol 53:673–684
Poelt J (1974) Systematik der Flechten. Bericht über die Jahre 1972 und 1973 mit einigen Nachtragen. Progr Bot 36:263–276
Poelt J, Vezda A (1981) Bestimmungsschlüssel europäischer Flechten. Erganzungsheft II. Bibl Lichenol 16, J. Cramer, Vaduz. 390 pp
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Prieto M, Wedin M (2013) Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS One 8(6) doi:10.1371/journal.pone.0065576
Purvis OW, Bailey EH, McLean J, Kasama T, Williamson BJ (2004) Uranium biosorption by the lichen Trapelia involuta at a uranium mine. Geomicrobiol J 21:159–167
Reeb V, Lutzoni F, Roux C (2004) Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina Fungi) with special emphasis on the lichen–forming Acarosporaceae and evolution of polyspory. Mol Phylogenet Evol 32:1036–1060
Rikkinen J (1995) What’s behind the pretty colours? A study on the photobiology of lichens. Bryobrothera 4:1–239
Ronquist F (2004) Bayesian inference of character evolution. Trends Ecol Evol 19:475–481
Ronquist F, Huelsenbeck J, Teslenko M (2011) Draft MrBayes version 3.2 Manual: Tutorials and Model Summaries. http://mrbayes.sourceforge.net/mb3.2_manual.pdf. Accessed 23 July 2014
Ronquist F, Teslenko M, van derMark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:1–4
Rothfels CJ, Larsson A, Kuo L-Y, Korall P, Chiou W-L, Prymer K-M (2012) Overcoming deep roots fast rates and short internodes to resolve the ancient rapid radiation of eupolypoid II ferns. Syst Biol 61:490–509
Schoch C, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny B, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JC, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister D, Peterson K, Gryzenhout M, Winngfield MJ, Aptroot A, Suh S-O, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman A, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O’Donnell K, Sipman H, Rogers JD, Shoemaker R, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW (2009) The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol 58:224–239
Schultz T, Churchill G (1999) The role of subjectivity in reconstructing ancestral character states: a Bayesian approach to unknown rates states and transformation asymmetries. Syst Biol 48:651–664
Solhaug KA, Gauslaa Y, Nybakken L, Bilger W (2002) UV-induction of sun-screening pigments in lichens. New Phytol 158:91–100
Spribille T, Muggia L (2013) Expanded taxon sampling disentangles evolutionary relationships and reveals a new family in Peltigerales (Lecanoromycetidae, Ascomycota). Fungal Divers 58:171–184
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stenroos SK, DePriest PT (1998) SSU rDNA phylogeny of cladoniiform lichens. Am J Bot 85:1548–1559
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577
Tehler A (1988) A cladistic outline of the Eumycota. Cladistics 4:227–277
Vězda A (1978) Neue oder wenig bekannte Flechten in der Tschechoslowakei. II. Folia Geobot Phytotax 13:397–4020
von Flotow J (1851) Über Psora privigna (Ach.) Fw. 1848. Bot Ztg (Berlin) 9:753–759
Wedin M, Döring H, Gilenstam G (2004) Saprotrophy and lichenization as options for the same fungal species on different substrata: environmental plasticity and fungal lifestyles in the Stictis-Conotrema complex. New Phytol 164:459–465
Wedin M, Wiklund E, Crewe A, Döring H, Ekman S, Nyberg A, Schmitt I, Lumbsch HT (2005) Phylogenetic relationships of Lecanoromycetes (Ascomycota) as revealed by analyses of mtSSU and nLSU rDNA sequence data. Mycol Res 109:159–172
Wedin M, Westberg M, Crewe AT, Tehler A, Purvis OW (2009) Species delimitation and evolution of metal bioaccumulation in the lichenized Acarospora smaragdula (Ascomycota Fungi) complex. Cladistics 25:161–172
Westberg M, Crewe AT, Purvis OW, Wedin M (2010[“2011”]) Silobia, a new genus for the Acarospora smaragdula complex (Ascomycota Acarosporales) and a revision of the group in Sweden. Lichenologist 43:7–25
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols. Academic Press, San Diego, pp 315–322
Yang Z (2008) Empirical evaluation of a prior for Bayesian phylogenetic inference. Philos Trans R Soc Lond B Biol Sci 363:4031–4039
Zahlbruckner A (1907) Lichenes, spezieller Teil. In Engler A, Prantl K. Die natürlichen Pflanzenfamilien I. Teil 1. Abteilung 49–249
Zoller S, Scheidegger C, Sperisen C (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31:511–516
Acknowledgments
This study was funded by grants to Martin Westberg by The Swedish Taxonomy Initiative (Svenska Artprojektet, administered by the Swedish Species Information Centre/ArtDatabanken) and was further supported by grants to Mats Wedin from the Swedish Research Council (VR 621-2009-5372, VR 621-2012-3990). The work of Kerry Knudsen was financially supported by the grant “Environmental aspects of sustainable development of society” 42900/1312/3166 from the Faculty of Environmental Sciences, Czech University of Life Sciences Prague. We are grateful to the staff at the Molecular Systematics Laboratory at the Swedish Museum of Natural History for laboratory assistance, in particular Jan Ohlson and Bodil Cronholm. Valerie Reeb kindly shared unpublished details from on her work on Acarosporaceae. The first author would finally like to thank Ulf Arup (LD), Philippe Clerc (G), Leif Tibell (UPS), Toni Berglund (Karlskoga) and members of the Swedish Lichen Society for assistance during field work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Tab. 1
(DOC 177 kb)
Suppl. Tab. 2
(DOC 49 kb)
Suppl. Tab. 3
(DOC 37 kb)
Suppl. Fig 1
Maximum likelihood best tree from the ITS dataset. Bootstrap values ≥ 70% –using 1000 Bootstrap replicates – are indicated over branches. Branch lengths are scaled to the expected number of nucleotide substitutions per site. (PDF 299 kb)
Suppl. Fig 2
Maximum likelihood best tree from the nSLU dataset. Bootstrap values ≥ 70% –using 1000 Bootstrap replicates – are indicated over branches. Branch lengths are scaled to the expected number of nucleotide substitutions per site. (PDF 268 kb)
Suppl. Fig 3
Maximum likelihood best tree from the mtSSU dataset. Bootstrap values ≥ 70% –using 1000 Bootstrap replicates – are indicated over branches. Branch lengths are scaled to the expected number of nucleotide substitutions per site. (PDF 268 kb)
Suppl. Fig 4
Maximum likelihood best tree from the BT dataset. Bootstrap values ≥ 70% –using 1000 Bootstrap replicates – are indicated over branches. Branch lengths are scaled to the expected number of nucleotide substitutions per site. (PDF 292 kb)
Rights and permissions
About this article
Cite this article
Westberg, M., Millanes, A.M., Knudsen, K. et al. Phylogeny of the Acarosporaceae (Lecanoromycetes, Ascomycota, Fungi) and the evolution of carbonized ascomata. Fungal Diversity 73, 145–158 (2015). https://doi.org/10.1007/s13225-015-0325-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-015-0325-x