Abstract
Osteosarcoma (OS) is the most common primary malignant bone tumor. However, the therapeutic results of the advanced cases at the first visit were still extremely poor. Therefore, more effective therapeutic options based on molecular profiling of OS are needed. In this study, we investigated the functions of endoplasmic reticulum (ER) stress activities in OS and elucidated whether ER stress inhibitors could exert antitumor effects. The expression of 84 key genes associated with unfolded protein response (UPR) was assessed in four OS cells (143B, MG63, U2OS and KHOS) by RT2 Profiler PCR Arrays. Based on results, we performed both siRNA and inhibitor assays focusing on IRE1α-XBP1 and PERK pathways. All OS cell lines showed resistance to PERK inhibitors. Furthermore, ATF4 and EIF2A inhibition by siRNA did not affect the survival of OS cell lines. On the other hand, IRE1α-XBP1 inhibition by toyocamycin suppressed OS cell growth (IC50: < 0.075 μM) and cell viability was suppressed in all OS cell lines by silencing XBP1 expression. The expression of XBP1s and XBP1u in OS cell lines and OS surgical samples were confirmed using qPCR. In MG63 and U2OS, toyocamycin decreased the expression level of XBP1s induced by tunicamycin. On the other hand, in 143B and KHOS, stimulation by toyocamycin did not clearly change the expression level of XBP1s induced by tunicamycin. However, morphological apoptotic changes and caspase activation were observed in these two cell lines. Inhibition of the IRE1α-XBP1s pathway is expected to be a promising new target for OS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor; it peaks during childhood/adolescence and after the age of 50 years. The standard protocol for the treatment of patients with OS was established more than 30 years ago (chemotherapy and surgical resection), and limited therapeutic progress has been made since then [1]. The therapeutic results of the advanced cases at the first visit were still extremely poor. Therefore, novel molecular targeted therapies and more effective therapeutic options based on molecular profiling of OS are needed.
Recently, studies have explored the therapeutic effects of targeting endoplasmic reticulum (ER) stress and unfolded protein response (UPR) using these inhibitors in several tumors [2, 3]. Our previous proteomic analyses demonstrated critical associations between ER stress response and malignant behaviors in Ewing’s sarcoma (ES) [2]. Furthermore, we found that IRE1α inhibitors exerted antitumor activity in ES [2]. However, the functional role of ER stress in OS has not been well elucidated. In this study, we investigated the functions of ER stress activities in OS and elucidated whether ER stress inhibitors could exert antitumor effects.
2 Material and methods
2.1 Cell lines
The 143B and MG63 cell lines were obtained from the American Type Culture Collection (ATCC). The KHOS and U2OS cell lines were provided by Dr. Melinda Merchant (National Cancer Institute, Bethesda, MD, USA). All cell lines were grown in Dulbecco’s modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin.
2.2 Array analyses of genes associated with UPR
The overall expression of 84 key genes associated with the UPR was determined with the RT2 Profiler PCR Arrays (PAHS-089Z; Qiagen, Venlo, The Netherlands), using an RT2 SYBR Green ROX qPCR Master Mix (Qiagen). Arrays were analyzed using mRNA from four OS cell lines. Thermal cycling was performed using ABI-7500Fast (Applied Biosystems, Foster, CA, USA) with initial denaturation at 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. The signal was acquired at 60 °C for each cycle. The cycle threshold (Ct) values obtained in quantification were used to calculate fold changes in mRNA abundance using the 2-∆∆Ct method.
2.3 RNA extraction and quantitative real-time PCR
RNA was extracted using RNeasy Plus Mini kit (Qiagen; Hilden, Germany). All quantitative real-time PCR (qPCR) was performed with TaqMan Fast Advanced Master Mix (Applied Biosystems) on an Applied Biosystems Step One Plus Real Time PCR System in accordance with standard protocols. qPCR was performed using predeveloped TaqMan assays (20 × Primer Probe mix; Applied Biosystems, CA, USA) for EIF2A (Assay ID Hs00230684_m1), ATF4 (Assay ID Hs00909569_g1), and GAPDH (Assay ID Hs02758991_g1). Custom qPCR sets were designed for XBP1s, XBP1u, and TATA-box binding protein (TBP) for separate quantifications. These primer and probe sequences were as follows: XBP1s (TaqMan custom probe: 5′-FAM-CTGGGCCTGCACCTGCTGCG-TAMRA-3′, primer sequences: 5′-CGCAGCAGGTGCAGGCCCAG-3′ and 5′- TTCTGGACAACTTGGACCCA-3′), XBP1u (TaqMan custom probe: 5′-FAM-AGCAGACCCGGCCACTGGCC -TAMRA-3′, primer sequences: 5’-GGCCAGTGGCCGGGTCTGCT-3’ and 5′-CTCAGACTACGTGCACCTC-3′), TBP (TaqMan custom probe: 5′-FAM-ACTGTTCTTCACTCTCTTGGCTCCTGTGCA-TAMRA-3′, primer sequences: 5′- GCATATTTTCTTGCTGCCAGTCT -3′ and 5′- ACCACGGCACTGATTTTCAGTT -3′). Plasmids for standard curves were generated by cloning cDNA fragments of XBP1s, XBP1u, and TBP into the pCRII TOPO vector (Invitrogen). The amounts of XBP1s and XBP1u relative to the housekeeping gene, TBP, were determined using the standard curve method. The amounts of other genes relative to the housekeeping gene, GAPDH, were determined using the comparative Ct method.
2.4 XBP1, eIF2a, and ATF4 siRNA knockdown in OS cell lines
For the knockdown expression studies, we used four cell lines (143B, MG63, KHOS, and U2OS). XBP1 siRNA knockdown was also performed using pre-designed XBP1 siRNA (sc-38627: Santa Cruz or siRNA negative control, Sigma-Aldrich), EIF2A siRNA (s38344, s38345: Silencer™ Select Pre-Designed siRNA or AM4611: Invitrogen™ Silencer™ Negative Control No. 1 siRNA), and ATF4 siRNA (s1702, s1703: Silencer™ Select Pre-Designed siRNA or AM4611: Invitrogen™ Silencer™ Negative Control No. 1 siRNA) using Lipofectamine™ RNAiMAX reagent (Thermo Fisher Scientific). After 72 h, RNA from each cell line was isolated, and its expression was validated using quantitative real-time PCR.
2.5 Cell proliferation with XBP1, eIF2a, and ATF4 siRNA knockdown
For knockdown proliferation studies with respect to XBP1, EIF2A, and ATF4, 2000 to 5000 OS cells were plated into 96-well plates on day 1. Transfection was performed on the same day with 25–50 nM of the siRNA reagents, as described above. After 72 h, the cell proliferation ability of OS cell lines was assessed using a Cell Counting Kit-8 (Dojindo Japan, Tokyo, Japan) and a microplate reader (SAFIRE, TECAN, Männedorf, Switzerland).
2.6 Growth inhibition assay
Toyocamycin (Tocris Bioscience, Bristol, UK) was used as an IRE1α-XBP1 pathway inhibitor. GSK2606414 (S7307, Selleck) and ISRIB (trans-isomer; S7400, Selleck) were used as PERK pathway inhibitors. OS cells were seeded into 96-well plates at a density of 3000–10000 cells/well. The next day, different concentrations of inhibitors or DMSO (as a vehicle control) were added to each well. After 72 h, the inhibitory effect of these inhibitors on the growth of OS cell lines was assessed using a Cell Counting Kit-8 (Dojindo Japan, Tokyo, Japan) and a microplate reader (SAFIRE, TECAN, Männedorf, Switzerland). The IC50 was calculated using GraphPad Prism software version 9.2.0 (GraphPad Software, Inc., CA, USA).
2.7 Apoptosis (caspase-3/7) assays
Because two OS cell lines (143B and KHOS) kept unexpectedly high-level expression of XBP1s even after Toyocamycin treatment while showing morphological apoptotic change, apoptotic assays were performed for these cell lines. These cells were plated into 96-well plates at a density of 5000 cells/well, and the next day, tunicamycin (TM) or DMSO (as a vehicle control) were added to each well. After 3 h and 6 h, apoptosis (caspase-3/7 activity) was measured using the Apo-ONE Homogeneous Caspase-3/7 Assay kit (no. G7791; Promega, Madison, WI, USA). Furthermore, apoptosis assay (caspase-3/7 activities) was performed at the following time points: after TM stimulation for 18 h, after TM stimulation for 6 h and subsequent toyocamycin stimulation (10–9 μM: minimum dose) for 12 h, and after DMSO (as a vehicle control) exposure for 18 h.
2.8 Statistical analysis
Statistical analyses were performed using GraphPad Prism® software version 9.2.0 P < 0.05 was considered significantly different.
3 Results
3.1 ER stress pathways are activated in OS cell lines
Three major signaling pathways in the ER stress response are inositol-requiring enzyme 1α (IRE1α), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6), all of which are involved in tumorigenesis [3,4,5]. We performed RT2 Profiler PCR Arrays to evaluate the expression of 84 key genes associated with the UPR (gene list in Supplementary Table 1). Among the three major signaling pathways in ER stress response, all four OS cell lines showed higher expression of PERK pathway genes, including ATF4 and EIF2A, followed by IRE1α pathway genes, including XBP1 (Supplementary Fig. 1A). ATF4 and EIF2A showed the highest and second highest expressions, respectively, among ER stress genes across all OS cell lines. In addition, XBP1 showed the third highest expression in three cell lines (143B, MG63, and U2OS), and the fifth highest expression in KHOS. Furthermore, stimulation with tunicamycin led to enhanced expression of PERK pathway genes, including ATF4, EIF2A, DDIT3, PPP1R15A, and DNAJC3 (Supplementary Fig. 1B). Furthermore, HSPA5, an upstream gene of the main UPR pathway, was also upregulated by tunicamycin stimulation. These findings indicated that tunicamycin stimulation enhanced the upstream UPR pathway gene and stimulated the PERK pathway among the three ER stress pathways. Based on these findings, we focused on the PERK and IRE1α pathways for further analysis.
3.2 OS cell lines showed resistance to the PERK inhibitors
Recently, two PERK inhibitors have been developed: GSK2606414 and ISRIB. GSK2606414 is an inhibitor of EIF2AK3 of the PERK pathway, while ISRIB is an inhibitor of EIF2A phosphorylation of the PERK pathway. The IC50 of GSK2606414 was shown to be 1.7 µM in ARPE-19 (normal epithelial cell line) treated with GSK2606414 for 72 h [6]. ISRIB alone has been reported to have poor antitumor effects on tumor cells [7]. In the present study, we verified the inhibitory effect of these inhibitors on OS cell lines. GSK2606414 did not show significant antitumor effects in any of the OS cell lines. All OS cell lines showed complete resistance to ISRIB (Fig. 1).
3.3 ATF4 and EIF2A inhibition by siRNA did not affect the survival of OS cell lines
To investigate the association between the PERK pathway and viability of OS cells, the inhibition of ATF4 and EIF2A was performed via siRNA-mediated knockdown of ATF4 and EIF2A in the four OS cell lines. qPCR confirmed a significant decrease in ATF4 and EIF2A mRNA levels in all OS cell lines (Supplementary Figs. 2A and 3A). In the cell proliferation assays, by silencing the expression of ATF4, cell viability was not significantly suppressed, except for U2OS (Supplementary Fig. 2B). Additionally, silencing EIF2A expression did not significantly suppress cell viability in any of the OS cell lines (Supplementary Fig. 3B).
3.4 IRE1α-XBP1 inhibition suppressed OS cell growth
Next, we examined the effect of IRE1α-XBP1 inhibition on OS cell lines. We had previously reported that toyocamycin showed the highest anti-tumor effect on Ewing’s sarcoma cells [2]. In Ewing sarcoma cell lines, it significantly and dose-dependently inhibited cell viability (IC50: 0.019 μM–0.050 μM) [2]. Toyocamycin also significantly and dose-dependently inhibited cell viability in OS cell lines as well (IC50: 0.027–0.072 μM) (Fig. 2). These findings suggest that Toyocamycin also has an inhibitory effect on OS cell lines.
3.5 The expression of XBP1s and XBP1u in OS cell lines and OS surgical samples
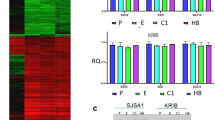
Our previous proteomic analyses demonstrated critical associations between the IRE1α-XBP1 pathway and malignant behaviors in Ewing sarcoma cells [2]. XBP1s and XBP1u expressions were analyzed in the four OS cell lines and eight clinical surgical materials. All OS cell lines showed the mRNA expression of XBP1s and XBP1u (Fig. 3A). MG63 cells had higher mRNA expression of XBP1s and XBP1u than the other three OS cell lines. Interestingly, all OS cell lines showed similar mRNA expression patterns of XBP1s/XBP1u. In OS surgical materials, all OS surgical materials showed mRNA expression of XBP1s and XBP1u, and XBP1s/XBP1u status also showed a trend similar to that of OS cell lines (Fig. 3B). XBP1s and XBP1u expression did not seem to be related to the chemotherapeutic state and histological type (Supplementary Table 2).
Expression of XBP1s and XBP1u in OS cell lines and clinical samples. A All OS cell lines show mRNA expression of XBP1s and XBP1u. MG63 has higher mRNA expression of XBP1s and XBP1u than the other three OS cell lines. B All OS surgical materials show mRNA expression of XBP1s and XBP1u; XBP1s/XBP1u status also shows a trend similar to that of the OS cell lines
3.6 The effects of silencing XBP1 on the viability of OS cell lines
To investigate the association between the IRE1α-XBP1 pathway and the survival of OS cell lines, inhibition of XBP1 by siRNA was performed in four OS cell lines. The knockdown of both XBP1s and XBP1u was confirmed using qPCR. In the cell proliferation assays, we also confirmed that cell viability was suppressed in all OS cell lines due to the silencing of XBP1 expression (Fig. 4A and B). These findings suggest a strong association between XBP1 expression and tumor proliferation in OS cells.
3.7 The effects of toyocamycin on the expression of XBP1s in OS cell lines
Toyocamycin is a selective IRE1α inhibitor that shows antitumor effects and induces apoptosis in cancer cells. Tunicamycin (TM) generally induces ER stress and enables the processing of XBP1u to XBP1s. Thus, we first stimulated OS cell lines with TM and evaluated XBP1s and XBP1u expression. TM stimulation (3 µg/ml) induced the expression of XBP1s in a time-dependent manner and suppressed the expression of XBP1u in all OS cell lines (Fig. 5). We next examined the inhibitory effects of toyocamycin on XBP1 cleavage after TM stimulation. In MG63 and U2OS cells, toyocamycin decreased the expression level of XBP1s induced by TM, and morphological apoptotic changes were not observed (Supplementary Fig. 5). On the other hand, in 143B and KHOS, stimulation by toyocamycin did not clearly change the expression level of XBP1s induced by TM. However, morphological apoptotic changes were observed in these two cell lines (Supplementary Fig. 5).
Expression level of XBP1s and XBP1u by stimulation with tunicamycin and toyocamycin. The expression of XBP1s is induced by tunicamycin (TM) stimulation (3 µg/ml) in a time-dependent manner, whereas the expression of XBP1u is suppressed in all OS cell lines. In MG63 and U2OS, the expression of XBP1s induced by TM is inhibited by toyocamycin. On the other hand, in 143B and KHOS, the expression level of XBP1s induced by TM is not changed clearly by stimulation with toyocamycin
3.8 Caspase-3/7 assay in OS cell liens
To verify the different effects of toyocamycin after TM stimulation on the two OS cell lines (143B and KHOS), we evaluated apoptotic activity using the caspase-3/7 assay. After TM stimulation for up to 6 h, caspase-3/7 activity was not evident in the OS cells, and morphological apoptotic change was not evident (Supplementary Figs. 4 and 5). Toyocamycin treatment at a low dose after TM stimulation elevated caspase-3/7 activity in two OS cell lines (143B and KHOS), and morphological apoptosis changes were evident (Supplementary Figs. 4 and 5). On the other hand, in MG63 and U2OS cells, morphological apoptosis changes were not evident (Supplementary Fig. 5) after TM stimulation for 6 h and toyocamycin treatment at a low dose after TM stimulation. These findings were consistent with the morphological changes observed in the OS cell lines following stimulation with toyocamycin.
4 Discussion
The endoplasmic reticulum (ER) is a major intracellular compartment involved in protein folding and maintenance of cell homeostasis [4, 8]. To maintain homeostasis in the ER, the amount of misfolded proteins is constantly monitored. The accumulation of misfolded proteins in the ER causes ER stress and initiates the unfolded protein response (UPR) to restore homeostasis [9]. However, under these long-term uncompensated ER stress conditions, the potential UPR makes it difficult to handle ER stress, leading to eventual cell apoptosis [8].
Tumor cells escape from ER stress by UPR, making the adjacent environment suitable for tumor survival and tumor growth [3, 10]. IRE1α, PERK, and ATF6 are three major signaling pathways involved in the ER stress response and tumorigenesis [3,4,5]. In bone and soft tissue tumors, our previous proteomic analyses demonstrated critical associations between ER stress response and malignant behaviors in Ewing’s sarcoma cells. Furthermore, we found that IRE1α inhibitors exerted antitumor activity in Ewing’s sarcoma cells [2]. To elucidate the potential of UPR as a therapeutic target in OS, we performed a comprehensive analysis of the ER stress response using RT2 Profiler PCR Arrays, and found high expression of PERK and IRE1α pathways-associated genes. Thus, we pursued these two pathways as possible therapeutic targets for OS. Regarding the relationship between PERK pathway and cancer, it has been pointed out that sustained PERK-EIF2A-ATF4 activation contributes to tumor progression and metastasis, and is ultimately associated with drug resistance [11], whereas under prolonged stress conditions of the ER, it leads to CHOP-induced apoptotic cell death [12]. In this study, blocking of the PERK pathway by siRNA and inhibitors did not affect the cell viability in OS, suggesting that the PERK pathway could not be a therapeutic target.
Several studies have revealed an association between the IRE1α pathway and malignant tumors, including apoptosis, cell differentiation, invasion, metastasis, and drug resistance [13]. XPB1 is a downstream transcriptional factor of the IRE1α pathway and plays an important role in cancer progression. It has been shown that the loss of XBP1 induces a terminal UPR that blocks proliferation and differentiation during mammary gland development [14]. In this study, knockdown of XBP1 strongly inhibited cell proliferation in all OS cell lines, which is consistent with a previous study showing the antitumor effect of XBP1 knockdown in two OS cell lines [15]. Functional analyses using IRE1α inhibitors have confirmed antitumor activity in several malignancies, including Ewing’s sarcoma cell lines, multiple myeloma, and pancreatic cancer [2, 16, 17]. Toyocamycin is an IRE1α inhibitor that exhibits antitumor effects by selectively inhibiting XBP1 mRNA splicing [17]. In all OS cell lines, Toyocamycin showed an antitumor effect similar to that in Ewing’s sarcoma cells [2]. These findings showed that blocking the IRE1α pathway could be a therapeutic target for OS.
Regarding XBP1 expression during TM/toyocamycin treatment, we found that TM stimulation induced XBP1s expression in all OS cell lines. Furthermore, we confirmed that XBP1s expression was decreased and XBP1u was increased after treatment with toyocamycin in two OS cell lines (U2OS and MG63). However, this switching of XBP1 expression after toyocamycin treatment was not clear in the other two OS cell lines (KHOS and 143B), and XBP1s expression remained at a high level. Interestingly, these two OS cell lines were not examined in a previous study showing anti-tumor effects on XBP1 blocking in OS [15]. Notably, these two OS cell lines showed morphological apoptotic changes, consistent with the finding that TM stimulation followed by low-dose toyocamycin treatment (12 h) increased apoptotic activity. Regarding the relationship between IRE1α pathway activation, including XBP1s overexpression and apoptosis, it has been known that activation of JNK (MAPK8) cooperates with p38 and induces apoptosis [5, 13]. However, in the comprehensive analysis of all OS cell lines stimulated with TM, MAPK8 expression was not enhanced (Supplementary Fig. 1B). Furthermore, it has been reported that sustained activation of XBP1 splicing induces apoptosis in normal tissues [18, 19]. Although it has not been reported whether sustained activation of XBP1 splicing induces apoptosis in tumor cells, we observed caspase activation in 143B and KHOS cells, after toyocamycin treatment at a low dose after TM stimulation, and morphological apoptotic changes were evident. Interestingly, TM treatment for 6 h followed by toyocamycin treatment for 12 h induced morphological apoptotic changes in 143B and KHOS with caspase activation, while high levels of XBP1s expression were preserved in these two cells as well as under TM stimulation. The reason for this paradoxical change in OS cells was unclear because high levels of XBP1s were preserved while morphological apoptotic changes occurred.
In Conclusion, we investigated the functions and malignant activities of ER stress response in OS, and further elucidated whether inhibitors of ER stress response had antitumor effects. Our findings demonstrated critical associations between ER stress response and malignant behavior in OS. Furthermore, we found that IRE1α inhibitors exerted antitumor activity in OS. As XBP1s expression was consistently observed in OS clinical samples and XBP1s expression was not related to the chemotherapeutic state, inhibition of this pathway is expected to be a new promising target for OS patients.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Change history
25 March 2022
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s12672-022-00481-6
References
Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, Harris MB, Healey J, Huvos A, Link M, Montebello J, Nadel H, Nieder M, Sato J, Siegal G, Weiner M, Wells R, Wold L, Womer R, Grier H. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23:2004–11. https://doi.org/10.1200/JCO.2005.06.031.
Tanabe Y, Suehara Y, Kohsaka S, Hayashi T, Akaike K, Mukaihara K, Kurihara T, Kim Y, Okubo T, Ishii M, Kazuno S, Kaneko K, Saito T. IRE1α-XBP1 inhibitors exerted anti-tumor activities in Ewing’s sarcoma. Oncotarget. 2018;9:14428–43. https://doi.org/10.18632/oncotarget.24467.
Oakes SA. Endoplasmic reticulum stress signaling in cancer cells. Am J Pathol. 2020;190:934–46. https://doi.org/10.1016/j.ajpath.2020.01.010.
Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. https://doi.org/10.1038/nrm3270.
Zhang T, Li N, Sun C, Jin Y, Sheng X. MYC and the unfolded protein response in cancer: synthetic lethal partners in crime? EMBO Mol Med. 2020;12: e11845. https://doi.org/10.15252/emmm.201911845.
Jiang X, Wei Y, Zhang T, Zhang Z, Qiu S, Zhou X, Zhang S. Effects of GSK2606414 on cell proliferation and endoplasmic reticulum stressassociated gene expression in retinal pigment epithelial cells. Mol Med Rep. 2017;15:3105–10. https://doi.org/10.3892/mmr.2017.6418.
Palam LR, Gore J, Craven KE, Wilson JL, Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 2015;6: e1913. https://doi.org/10.1038/cddis.2015.264.
Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–30. https://doi.org/10.1038/nrd2755.
Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69:169–81. https://doi.org/10.1016/j.molcel.2017.06.017.
Yadav RK, Chae SW, Kim HR, Chae HJ. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19:75–88. https://doi.org/10.15430/JCP.2014.19.2.75.
Bu Y, Diehl JA. PERK integrates oncogenic signaling and cell survival during cancer development. J Cell Physiol. 2016;231:2088–96. https://doi.org/10.1002/jcp.25336.
Rozpędek W, Pytel D, Mucha B, Leszczyńska H, Diehl JA, Majsterek I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med. 2016;16:533–44. https://doi.org/10.2174/1566524016666160523143937.
Chen S, Chen J, Hua X, Sun Y, Cui R, Sha J, Zhu X. The emerging role of XBP1 in cancer. Biomed Pharmacother. 2020;127: 110069. https://doi.org/10.1016/j.biopha.2020.110069.
Hasegawa D, Calvo V, Avivar-Valderas A, Lade A, Chou HI, Lee YA, Farias EF, Aguirre-Ghiso JA, Friedman SL. Epithelial Xbp1 is required for cellular proliferation and differentiation during mammary gland development. Mol Cell Biol. 2015;35:1543–56. https://doi.org/10.1128/MCB.00136-15.
Yang J, Cheng D, Zhou S, Zhu B, Hu T, Yang Q. Overexpression of X-Box binding protein 1 (XBP1) correlates to poor prognosis and up-regulation of PI3K/mTOR in human osteosarcoma. Int J Mol Sci. 2015;16:28635–46. https://doi.org/10.3390/ijms161226123.
Chien W, Ding L-W, Sun Q-Y, Torres-Fernandez LA, Tan SZ, Xiao J, Lim SL, Garg M, Lee KL, Kitajima S, Takao S, Leong WZ, Sun H, Tokatly I, Poellinger L, Gery S, Koeffler PH. Selective inhibition of unfolded protein response induces apoptosis in pancreatic cancer cells. Oncotarget. 2014;5:4881–94. https://doi.org/10.18632/oncotarget.2051.
Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, Narita T, Masaki A, Ito A, Ding J, Kusumoto S, Ishida T, Komatsu H, Shiotsu Y, Ueda R, Iwawaki T, Imoto M, Iida S. Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer J. 2012;2: e79. https://doi.org/10.1038/bcj.2012.26.
Allagnat F, Christulia F, Ortis F, Pirot P, Lortz S, Lenzen S, Eizirik DL, Cardozo AK. Sustained production of spliced X-box binding protein 1 (XBP1) induces pancreatic beta cell dysfunction and apoptosis. Diabetologia. 2010;53:1120–30. https://doi.org/10.1007/s00125-010-1699-7.
Zeng L, Zampetaki A, Margariti A, Pepe AE, Alam S, Martin D, Xiao Q, Wang W, Jin Z-G, Cockerill G, Mori K, Li Y-SJ, Hu Y, Chien S, Xu Q. Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc Natl Acad Sci USA. 2009;106:8326–31. https://doi.org/10.1073/pnas.0903197106.
Funding
This study was supported by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) KAKENHI (JSPS: Grant Numbers #19H03789 and #19K22694 to Y.S., #19K16753 to K.A., #18K15329 to T.O., #20K22963 to T.K., and #20K07415 and #17K08730 to T.S.).
Author information
Authors and Affiliations
Contributions
KS: Investigation, methodology, formal analysis, data curation, writing—original draft, writing—review and editing. TS: Conceptualization, methodology, formal analysis, data curation, funding acquisition, wrinting—original draft, writing—review and editing, supervision, project administration. TK: Methodology, formal analysis, data curation, funding acquisition. NH: Methodology, formal analysis, data curation. KS: Methodology, formal analysis, data curation. DK: Resources, methodology, formal analysis, data curation. KA: Resources, methodology, formal analysis, data curation, funding acquisition. TO: Resources, methodology, formal analysis, data curation, funding acquisition. TH: Methodology, formal analysis, data curation, writing—review and editing. TT: Supervision, resources, data curation. TY: Supervision, data curation, writing—review and editing. MI: Supervision, data curation, writing—review and editing. YS: Conceptualization, methodology, formal analysis, data curation, project administration, funding acquisition, writing—review and editing, supervision. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was reviewed and approved by Juntendo University School of Medicine Institutional Review Board (#21-079).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s12672-022-00481-6
Supplementary Information
12672_2021_453_MOESM1_ESM.xlsx
Additional file 1: Table S1. List of the 84 key genes associated with the UPR (unfolded protein response) on RT2 Profiler PCR Arrays. (XLSX 15 KB)
12672_2021_453_MOESM3_ESM.pptx
Additional file 3: Figure S1A-1. Gene expression data of the 84 key genes associated with the UPR without any stimulation (143B and MG63). Green: PERK pathway; Yellow: IRE1α pathway; Blue: ATF6 pathway. Figure S1A-2. Gene expression data of the 84 key genes associated with the UPR without any stimulation (KHOS and U2OS). Green: PERK pathway; Yellow: IRE1α pathway; Blue: ATF6 pathway. Figure S1B: ER stress response analysis using the RT2 Profiler PCR Arrays after TM stimulation shows a high expression of DDIT3 (CHOP) followed by EIL2A and ATF4. XBP1s and XBP1u were not included in this array (listed as XBP1; thus, could not be distinguished from each other), and were, therefore, separately examined using qPCR. Figure S2: Cell viability by ATF4 knockdown in OS cell lines. ATF4 siRNA knockdown in four OS cell lines (143B, MG63, KHOS, and U2OS) was performed to examine the associations between ATF4 expression and cell viability. A Suppression of ATF4 expression evaluated using qPCR. The expression of ATF4 in each OS cell line is suppressed by all ATF4 siRNAs. B The cell viability in OS cell lines is not affected by ATF4 siRNA knockdown. Figure S3 Cell viability following EIF2A knockdown in OS cell lines. EIF2A siRNA knockdown in four OS cell lines (143 B, MG63, KHOS, and U2OS) was performed to examine the association between EIF2A expression and cell viability. A Suppression of EIF2A expression is evaluated using qPCR. The expression of EIF2A in each OS cell line is suppressed by all EIF2A siRNAs. B The cell viability in OS cell lines is not affected by EIF2A siRNA knockdown. Figure S4. Caspase-3/7 assay in OS cell liens. Caspase-3/7 activity is not evident in OS cells after TM stimulation for up to 6 h. However, it is activated by prolonged stimulation (18 h) in KHOS cell. Caspase-3/7 activity in 143B and KHOS cells is elevated by toyocamycin treatment after TM stimulation. Figure S5. Morphological apoptotic changes in OS cell liens. In MG63 and U2OS cells, morphological apoptosis changes are not evident after TM stimulation for up to 6 h and toyocamycin treatment after TM stimulation. On the other hand, in 143B and KHOS cells, morphological apoptosis changes are evident after low dose toyocamycin treatment after TM stimulation. (PPTX 4759 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sasa, K., Saito, T., Kurihara, T. et al. RETRACTED ARTICLE: IRE1α-XBP1 but not PERK inhibition exerts anti-tumor activity in osteosarcoma. Discov Onc 12, 57 (2021). https://doi.org/10.1007/s12672-021-00453-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-021-00453-2