Abstract
Hospital information systems (HIS) are service-oriented systems that focus on payment for medical services. Because all HIS coding for diseases and clinical processes are payment-oriented, they may differ from clinicians’ concepts of diseases and processes. HIS in large-scale hospitals in Japan utilize Diagnostic Procedure Combination (DPC) codes, a disease-coding system that focuses on the use of medical resources. Although DPC codes are very precise for diseases requiring surgery, such as cataracts and lung cancer, classification codes for diseases that do not require surgery, such as cerebral infarction, are less precise, with a single category often covering many subtypes with different clinical courses. This paper proposes a preprocessing method that splits DPC codes into subgroups prior to the application of dual clustering-based clinical pathway mining. This method applies expectation–maximization (EM) clustering to the length of patient stay in the hospital using Akaike Information Criteria (AIC) to select the number of clusters. A dual mining method is subsequently applied to the datasets of subgroups and the meanings of subtype clusters are explored using a text mining method. The proposed method was evaluated using datasets from an HIS at Shimane University hospital as preprocessing for clinical pathway mining. The experimental results showed that the proposed method correctly generated subgroups from the more generalized DPC codes and that the clinical pathways identified after this preprocessing capture the characteristics of processes in real clinical settings.








Similar content being viewed by others
Notes
The reviewer asked the authors about the true values for a clinical pathway. From the dataset, the true value is regarded as whether the nursing activity was or was not performed frequently. If performed frequently, the instruction was selected as an element of the clinical pathway.
In statistics, \(\theta\) usually denotes the statistical distribution of a parameter. For example, a normally distributed parameter can be represented as the mean (\(\theta _1=\mu\)) and variance (\(\theta _2=\sigma\)) [2].
AIC and Log-likelihood were calculated using R-package mixtools [3]. Unfortunately, mixtools could not determine several statistical parameters, such as dispersion of AIC.
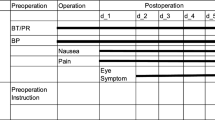
Because most patients with cerebral infarction do not undergo surgery, the preoperative period (such as day − 1) is not included.
Although the darch package was used, darch has since been removed from R package (CRAN). Check the github (https://github.com/maddin79/darch) for this package.
References
Akaike, H. (1973). Information theory and an extension of the maximum likelihood principle. In: B.N. Petrov, & F. Caski (Eds.) Proceedings of the 2nd International Symposium on Information Theory (pp. 267–281). Akadimiai Kiado.
Analytics-Toolkit.com .(2021). What does “thera” mean ? https://www.analytics-toolkit.com/glossary/theta/
Benaglia, T., Chauveau, D., Hunter, D. R., & Young, D. (2009). mixtools: An r package for analyzing finite mixture models. Journal of Statistical Software, 32(6), 1–29.
Esteva, A., Robicquet, A., Ramsundar, B., Kuleshov, V., DePristo, M., Chou, K., et al. (2019). A guide to deep learning in healthcare. Nature Medicine, 25(1), 24–50.
Hyde, E., & Murphy, B. (2012). Computerized clinical pathways (care plans): piloting a strategy to enhance quality patient care. Clinical Nurse Specialist, 26(4), 277–282.
Igakutsushinsha. (2020). Quick Reference of DPC points (in Japanese). Igakutsushinsha.
Ishida, M. (2016). Rmecab. http://rmecab.jp/wiki/index.php?RMeCabFunctions.
Iwata, H., Hirano, S., & Tsumoto, S. (2014). Construction of clinical pathway based on similarity-based mining in hospital information system. In Proceedings of the Second International Conference on Information Technology and Quantitative Management, ITQM 2014, National Research University Higher School of Economics (HSE), Moscow, Russia, June 3-5, 2014 (pp. 1107–1115). https://doi.org/10.1016/j.procs.2014.05.366.
Iwata, H., Hirano, S., & Tsumoto, S. (2015). Maintenance and discovery of domain knowledge for nursing care using data in hospital information system. Fundam. Inform.,137(2), 237–252. https://doi.org/10.3233/FI-2015-1177. http://dx.doi.org/10.3233/FI-2015-1177
Kim, J.H. (2009). Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Computational Statistics & Data Analysis,53(11), 3735–3745. https://doi.org/10.1016/j.csda.2009.04.009.
Liaw, A., & Wiener, M. (2002). Classification and regression by randomforest. R News, 2(3), 18–22. http://CRAN.R-project.org/doc/Rnews/.
McLachlan, G. J., & Peel, D. (2000). Finite Mixture Models. New York: Wiley.
Melton, G., McDonald, C.J., Tang, P.C., & Hripcsak, G. (2014). Elecronic Health Records (4th edn., chap. 16, 2014). Springer.
Motoda, H. (Ed.). (2002). Active Mining. No. 79 in Frontiers in Artificial Intelligence and Applications. IOS Press.
Organization, W.H. (1993). ICD-10. ICD-10 / World Health Organization. World Health Organization.
Schwarz, G. (1978). Estimating the dimension of a model. The Annals of Statistics, 461–464.
Topol, E. (2019). High-performance medicine: the convergence of human and artificial intelligence. Nature Medicine, 25(1), 44–56.
Tsumoto, S., Hirano, S., & Iwata, H. (2016). Construction of clinical pathway from histories of clinical actions in hospital information system. In 2016 IEEE International Conference on Big Data, BigData 2016, Washington DC, USA, December 5-8, 2016 (pp. 1972–1981). https://doi.org/10.1109/BigData.2016.7840819.
Tsumoto, S., Iwata, H., Hirano, S., & Tsumoto, Y. (2014). Similarity-based behavior and process mining of medical practices. Future Generation Computer Systems, 33, 21–31. https://doi.org/10.1016/j.future.2013.10.014.
Tsumoto, S., Kimura, T., & Hirano, S. (2021). Determinaion of diseases from discharge summaries—a text mining approach. Review of Socionetwork Strategies, 15, 49–66.
Tsumoto, S., Kimura, T., Iwata, H., & Hirano, S. (2017). Construction of discharge summaries classifier. In 2017 IEEE International Conference on Healthcare Informatics, ICHI 2017, Park City, UT, USA, August 23-26, 2017 (pp. 74–82). IEEE. https://doi.org/10.1109/ICHI.2017.92.
Tsumoto, S., Kimura, T., Iwata, H., & Hirano, S. (2021). Mining clinical pathways using dual clustering. Review of Socionetwork Strategies, 15, 1–21.
Tsumoto, S., Yamaguchi, T., Numao, M., & Motoda, H. (Eds.). (2005). Active Mining, Second International Workshop, AM 2003, Maebashi, Japan, October 28, 2003, Revised Selected Papers. Lecture Notes in Computer Science (vol. 3430). Springer.
Tsumoto, Y., Iwata, H., Hirano, S., & Tsumoto, S. (2015). Construction of clinical pathway using dual clustering. Neuroscience and Biomedical Engineering, 3, 49–56.
Ward, M., Vartak, S., Schwichtenberg, T., & Wakefield, D. (2011). Nurses’ perceptions of how clinical information system implementation affects workflow and patient care. Computers, Informatics, Nursing, 29(9), 502–511.
Acknowledgements
This research was supported by Grant-in-Aid for Scientific Research (B) 18H03289 from Japan Society for the Promotion of Science(JSPS). The authors thanks Dominik Ślȩzak, Tzung-Pei Hong and Weiping Ding for their useful comments on information granulation (granular comptuing). The authors also thanks the reviewers for insightful comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author states that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research was supported by Grant-in-Aid for Scientific Research (B) 18H03289 from Japan Society for the Promotion of Science (JSPS).
Rights and permissions
About this article
Cite this article
Tsumoto, S., Kimura, T. & Hirano, S. Expectation–Maximization (EM) Clustering as a Preprocessing Method for Clinical Pathway Mining. Rev Socionetwork Strat 16, 25–52 (2022). https://doi.org/10.1007/s12626-021-00100-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12626-021-00100-w