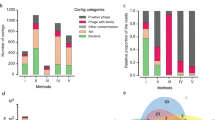
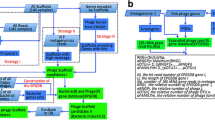
Abstract
CrAss-like phages are a diverse group of bacteriophages genetically similar to the prototypical crAssphage (p-crAssphage), which was discovered in the human gut microbiome through a metagenomics approach. It was identified as a ubiquitous and highly abundant bacteriophage group in the gut microbiome. Initial co-occurrence analysis postulated Bacteroides spp. as the prospective bacterial host. Subsequent studies have confirmed multiple host species under Phylum Bacteroidetes and some Firmicutes. Detection of crAss-like phages in sewage-contaminated environmental water and robust correlation with enteric viruses and bacteria has culminated in their adoption as a microbial source tracking (MST) marker. Polymerase chain reaction (PCR) and real-time PCR assays have been developed utilizing the conserved genes in the p-crAssphage genome to detect human fecal contamination of different water sources, with high specificity. Numerous investigations have examined the implications of crAss-like phages in diverse disease conditions, including ulcerative colitis, obesity and metabolic syndrome, autism spectrum disorders, rheumatoid arthritis, atopic eczema, and other autoimmune disorders. These studies have unveiled associations between certain diseases and diminished abundance and diversity of crAss-like phages. This review offers insights into the diverse aspects of research on crAss-like phages, including their discovery, genomic characteristics, structure, taxonomy, isolation, molecular detection, application as an MST marker, and role as a gut microbiome modulator with consequential health implications.

Similar content being viewed by others
Data Availability
All data were obtained from publicly available information.
Code Availability
Not applicable.
References
Ahmed, W., Bivins, A., Payyappat, S., Cassidy, M., Harrison, N., & Besley, C. (2022). Distribution of human fecal marker genes and their association with pathogenic viruses in untreated wastewater determined using quantitative PCR. Water Research, 226, 119093.
Ahmed, W., Gyawali, P., Hamilton, K. A., Joshi, S., Aster, D., Donner, E., Simpson, S. L., & Symonds, E. M. (2021). Antibiotic resistance and sewage-associated marker genes in untreated sewage and a river characterized during baseflow and stormflow. Frontiers in Microbiology, 12, 632850.
Ahmed, W., Lobos, A., Senkbeil, J., Peraud, J., Gallard, J., & Harwood, V. J. (2018a). Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Research, 131, 142–150.
Ahmed, W., Payyappat, S., Cassidy, M., & Besley, C. (2019a). A duplex PCR assay for the simultaneous quantification of Bacteroides HF183 and crAssphage CPQ_056 marker genes in untreated sewage and stormwater. Environment International, 126, 252–259.
Ahmed, W., Payyappat, S., Cassidy, M., & Besley, C. (2019b). Enhanced insights from human and animal host-associated molecular marker genes in a freshwater lake receiving wet weather overflows. Scientific Reports, 9(1), 1–13.
Ahmed, W., Payyappat, S., Cassidy, M., Besley, C., & Power, K. (2018b). Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Research, 145, 769–778.
Ahmed, W., Payyappat, S., Cassidy, M., Harrison, N., & Besley, C. (2023a). Microbial source tracking of untreated human wastewater and animal scats in urbanized estuarine waters. Science of the Total Environment, 877, 162764.
Ahmed, W., Payyappat, S., Cassidy, M., Harrison, N., & Besley, C. (2023). Reduction of human fecal markers and enteric viruses in Sydney estuarine waters receiving wet weather overflows. Science of The Total Environment, 896, 165008.
Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., & Wang, J. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host & Microbe, 17(5), 690–703.
Ballesté, E., Pascual-Benito, M., Martín-Díaz, J., Blanch, A. R., Lucena, F., Muniesa, M., & García-Aljaro, C. (2019). Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water research, 155, 233–244.
Bayfield, O. W., Shkoporov, A. N., Yutin, N., Khokhlova, E. V., Smith, J. L., Hawkins, D. E., & Antson, A. A. (2023). Structural atlas of a human gut crassvirus. Nature. https://doi.org/10.1038/s41586-023-06019-2
Boix-Amorós, A., Monaco, H., Sambataro, E., & Clemente, J. C. (2022). Novel technologies to characterize and engineer the microbiome in inflammatory bowel disease. Gut Microbes, 14(1), 2107866.
Cadwell, K. (2015). The virome in host health and disease. Immunity, 42(5), 805–813.
Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D., & Lawley, T. D. (2021). Massive expansion of human gut bacteriophage diversity. Cell, 184(4), 1098–1109.
Cannon, J. L., Park, G. W., Anderson, B., Leone, C., Chao, M., Vinjé, J., & Fraser, A. M. (2022). Hygienic monitoring in long-term care facilities using ATP, crAssphage, and human noroviruses to direct environmental surface cleaning. American Journal of Infection Control, 50(3), 289–294.
Carding, S. R., Davis, N., & Hoyles, L. J. A. P. (2017). The human intestinal virome in health and disease. Alimentary Pharmacology & Therapeutics, 46(9), 800–815.
Cervantes-Echeverría, M., Gallardo-Becerra, L., Cornejo-Granados, F., & Ochoa-Leyva, A. (2023). The two-faced role of crassphage subfamilies in obesity and metabolic syndrome: Between good and evil. Genes, 14(1), 139.
Chu, Y., Meng, Q., Yu, J., Zhang, J., Chen, J., & Kang, Y. (2023). Strain-level dynamics reveal regulatory roles in atopic eczema by gut bacterial phages. Microbiology Spectrum, 11(2), e04551-e4622.
Cinek, O., Kramna, L., Lin, J., Oikarinen, S., Kolarova, K., Ilonen, J., & Hyöty, H. (2017). Imbalance of bacteriome profiles within the finnish diabetes prediction and prevention study: Parallel use of 16S profiling and virome sequencing in stool samples from children with islet autoimmunity and matched controls. Pediatric Diabetes, 18(7), 588–598.
Clooney, A. G., Sutton, T. D., Shkoporov, A. N., Holohan, R. K., Daly, K. M., O’Regan, O., & Hill, C. (2019). Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host & Microbe, 26(6), 764–778.
Crank, K., Li, X., North, D., Ferraro, G. B., Iaconelli, M., Mancini, P., & Bibby, K. (2020). CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Research, 184, 116161.
Cresci, G. A., & Izzo, K. (2019). Gut microbiome. In M. L. Corrigan, K. Roberts, & E. Steiger (Eds.), Adult short bowel syndrome: Nutritional, medical, and surgical management (pp. 45–54). Academic Press.
Cuevas-Ferrando, E., Pérez-Cataluña, A., Falcó, I., Randazzo, W., & Sánchez, G. (2022). Monitoring human viral pathogens reveals potential hazard for treated wastewater discharge or reuse. Frontiers in Microbiology, 13, 779.
Dias, E., Ebdon, J., & Taylor, H. (2018). The application of bacteriophages as novel indicators of viral pathogens in wastewater treatment systems. Water Research, 129, 172–179.
Dominianni, C., Sinha, R., Goedert, J. J., Pei, Z., Yang, L., Hayes, R. B., & Ahn, J. (2015). Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS ONE, 10(4), e0124599.
Draper, L. A., Ryan, F. J., Smith, M. K., Jalanka, J., Mattila, E., Arkkila, P. A., & Hill, C. (2018). Long-term colonization with donor bacteriophages following successful faecal microbial transplantation. Microbiome, 6(1), 1–9.
Dutilh, B. E., Cassman, N., McNair, K., Sanchez, S. E., Silva, G. G., Boling, L., & Edwards, R. A. (2014). A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nature Communications, 5(1), 4498.
Edwards, R. A., Vega, A. A., Norman, H. M., Ohaeri, M., Levi, K., Dinsdale, E. A., & Dutilh, B. E. (2019). Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nature Microbiology, 4(10), 1727–1736.
Farkas, K., Adriaenssens, E. M., Walker, D. I., McDonald, J. E., Malham, S. K., & Jones, D. L. (2019). Critical evaluation of CrAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food and Environmental Virology, 11, 113–119.
García-Aljaro, C., Ballesté, E., Muniesa, M., & Jofre, J. (2017). Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microbial Biotechnology, 10(6), 1775–1780.
Gerrity, D., Papp, K., Dickenson, E., Ejjada, M., Marti, E., Quinones, O., & Trenholm, R. A. (2022). Characterizing the chemical and microbial fingerprint of unsheltered homelessness in an urban watershed. Science of The Total Environment, 840, 156714.
Gorvitovskaia, A., Holmes, S. P., & Huse, S. M. (2016). Interpreting prevotella and bacteroides as biomarkers of diet and lifestyle. Microbiome, 4, 15.
Green, H., Wilder, M., Collins, M., Fenty, A., Gentile, K., Kmush, B. L., & Larsen, D. A. (2020). Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. MedRxiv. https://doi.org/10.1101/2020.05.21.20109181
Gregory, A. C., Zablocki, O., Zayed, A. A., Howell, A., Bolduc, B., & Sullivan, M. B. (2020). The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host & Microbe, 28(5), 724–740.
Guerin, E., Shkoporov, A., Stockdale, S. R., Clooney, A. G., Ryan, F. J., Sutton, T. D., & Hill, C. (2018). Biology and taxonomy of crAss-like bacteriophages, the most abundant virus in the human gut. Cell Host & Microbe, 24(5), 653–664.
Guerin, E., Shkoporov, A. N., Stockdale, S. R., Comas, J. C., Khokhlova, E. V., Clooney, A. G., & Hill, C. (2021). Isolation and characterization of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens. Microbiome, 9, 1–21.
Gulyaeva, A., Garmaeva, S., Ruigrok, R. A., Wang, D., Riksen, N. P., Netea, M. G., & Zhernakova, A. (2022). Discovery, diversity, and functional associations of crAss-like phages in human gut metagenomes from four Dutch cohorts. Cell Reports. https://doi.org/10.1016/j.celrep.2021.110204
Gyawali, P., Devane, M., Scholes, P., & Hewitt, J. (2021). Application of crAssphage, F-RNA phage and pepper mild mottle virus as indicators of human faecal and norovirus contamination in shellfish. Science of the Total Environment, 783, 146848.
Hamza, I. A., & Abd-Elmaksoud, S. (2023). Applicability of crAssphage as a performance indicator for viral reduction during activated sludge wastewater treatment. Environmental Science and Pollution Research, 30(17), 50723–50731.
Holm, R. H., Nagarkar, M., Yeager, R. A., Talley, D., Chaney, A. C., Rai, J. P., & Smith, T. (2022). Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods Kentucky. FEMS Microbes, 3, xtac003.
Honap, T. P., Sankaranarayanan, K., Schnorr, S. L., Ozga, A. T., Warinner, C., & Lewis, C. M., Jr. (2020). Biogeographic study of human gut-associated crAssphage suggests impacts from industrialization and recent expansion. PLoS ONE, 15(1), e0226930.
Hryckowian, A. J., Merrill, B. D., Porter, N. T., Van Treuren, W., Nelson, E. J., Garlena, R. A., & Sonnenburg, J. L. (2020). Bacteroides thetaiotaomicron-infecting bacteriophage isolates inform sequence-based host range predictions. Cell Host & Microbe, 28(3), 371–379.
Jennings, W. C., Gálvez-Arango, E., Prieto, A. L., & Boehm, A. B. (2020). CrAssphage for fecal source tracking in Chile: Covariation with norovirus, HF183, and bacterial indicators. Water Research X, 9, 100071.
Karkman, A., Pärnänen, K., & Larsson, D. J. (2019). Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nature Communications, 10(1), 80.
Kongprajug, A., Chyerochana, N., Rattanakul, S., Denpetkul, T., Sangkaew, W., Somnark, P., & Sirikanchana, K. (2021). Integrated analyses of fecal indicator bacteria, microbial source tracking markers, and pathogens for Southeast Asian beach water quality assessment. Water Research, 203, 117479.
Kongprajug, A., Mongkolsuk, S., & Sirikanchana, K. (2019). CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Environmental Science & Technology Letters, 6(3), 159–164.
Korajkic, A., Kelleher, J., Shanks, O. C., Herrmann, M. P., & McMinn, B. R. (2022). Effectiveness of two wastewater disinfection strategies for the removal of fecal indicator bacteria, bacteriophage, and enteric viral pathogens concentrated using dead-end hollow fiber ultrafiltration (D-HFUF). Science of the Total Environment, 831, 154861.
Korajkic, A., McMinn, B., Herrmann, M. P., Sivaganesan, M., Kelty, C. A., Clinton, P., & Shanks, O. C. (2020). Viral and bacterial fecal indicators in untreated wastewater across the contiguous United States exhibit geospatial trends. Applied and Environmental Microbiology, 86(8), e02967.
Lee, S. Y., Yang, J., & Lee, J. H. (2023). Improvement of crAssphage detection/quantification method and its extensive application for food safety. Frontiers in Microbiology, 14, 1185788.
Li, M., Wang, B., Zhang, M., Rantalainen, M., Wang, S., Zhou, H., & Zhao, L. (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences, 105(6), 2117–2122.
Li, W., Liu, Z., Hu, B., & Zhu, L. (2021a). Co-occurrence of crAssphage and antibiotic resistance genes in agricultural soils of the Yangtze River Delta, China. Environment International, 156, 106620.
Li, Y., Gordon, E., Shean, R. C., Idle, A., Deng, X., Greninger, A. L., & Delwart, E. (2021b). CrAssphage and its bacterial host in cat feces. Scientific Reports, 11(1), 815.
Liang, Y., Jin, X., Huang, Y., & Chen, S. (2018). Development and application of a real-time polymerase chain reaction assay for detection of a novel gut bacteriophage (crAssphage). Journal of Medical Virology, 90(3), 464–468.
Liang, Y. Y., Zhang, W., Tong, Y. G., & Chen, S. P. (2016). crAssphage is not associated with diarrhoea and has high genetic diversity. Epidemiology & Infection, 144(16), 3549–3553.
Liu, Z., Lin, Y., Ge, Y., Zhu, Z., Yuan, J., Yin, Q., & Hu, M. (2023). Meta-analysis of microbial source tracking for the identification of fecal contamination in aquatic environments based on data-mining. Journal of Environmental Management, 345, 118800.
Makkaew, P., Kongprajug, A., Chyerochana, N., Sresung, M., Precha, N., Mongkolsuk, S., & Sirikanchana, K. (2021). Persisting antibiotic resistance gene pollution and its association with human sewage sources in tropical marine beach waters. International Journal of Hygiene and Environmental Health, 238, 113859.
Malla, B., Ghaju Shrestha, R., Tandukar, S., Sherchand, J. B., & Haramoto, E. (2019a). Performance evaluation of human-specific viral markers and application of pepper mild mottle virus and crAssphage to environmental water samples as fecal pollution markers in the Kathmandu Valley, Nepal. Food and Environmental Virology, 11, 274–287.
Malla, B., Makise, K., Nakaya, K., Mochizuki, T., Yamada, T., & Haramoto, E. (2019b). Evaluation of human-and animal-specific viral markers and application of CrAssphage, pepper mild mottle virus, and tobacco mosaic virus as potential fecal pollution markers to river water in Japan. Food and Environmental Virology, 11, 446–452.
Manrique, P., Bolduc, B., Walk, S. T., van der Oost, J., de Vos, W. M., & Young, M. J. (2016). Healthy human gut phageome. Proceedings of the National Academy of Sciences, 113(37), 10400–10405.
McCann, A., Ryan, F. J., Stockdale, S. R., Dalmasso, M., Blake, T., Ryan, C. A., & Hill, C. (2018). Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ, 6, e4694.
Meuchi, Y., Nakada, M., Kuroda, K., Hanamoto, S., & Hata, A. (2023). Applicability of F-specific bacteriophage subgroups, PMMoV and crAssphage as indicators of source specific fecal contamination and viral inactivation in rivers in Japan. PLoS ONE, 18(7), e0288454.
Morrison, C. M., Betancourt, W. Q., Quintanar, D. R., Lopez, G. U., Pepper, I. L., & Gerba, C. P. (2020). Potential indicators of virus transport and removal during soil aquifer treatment of treated wastewater effluent. Water Research, 177, 115812.
Mueller, S., Saunier, K., Hanisch, C., Norin, E., Alm, L., Midtvedt, T., & Blaut, M. (2006). Differences in fecal microbiota in different European study populations in relation to age, gender, and country: A cross-sectional study. Applied and Environmental Microbiology, 72(2), 1027.
Nam, S. J., Kim, D. W., Lee, S. H., & Koo, O. K. (2023). Assessment of microbial source tracking marker and fecal indicator bacteria on food-contact surfaces in school cafeterias. Journal of Food Protection, 86(2), 100035.
Nguyen, K., Smith, S., Roundtree, A., Feistel, D. J., Kirby, A. E., Levy, K., & Mattioli, M. C. (2022). Fecal indicators and antibiotic resistance genes exhibit diurnal trends in the Chattahoochee River: Implications for water quality monitoring. Frontiers in Microbiology, 13, 1029176.
Nguyen, N. T., Liu, M., Katayama, H., Takemura, T., & Kasuga, I. (2021). Association of the colistin resistance gene mcr-1 with faecal pollution in water environments in Hanoi, Vietnam. Letters in Applied Microbiology, 72(3), 275–282.
Papp, K., Moser, D., & Gerrity, D. (2020). Viral surrogates in potable reuse applications: Evaluation of a membrane bioreactor and full advanced treatment. Journal of Environmental Engineering, 146(2), 04019103.
Papudeshi, B., Vega, A. A., Souza, C., Giles, S. K., Mallawaarachchi, V., Roach, M. J., & Edwards, R. A. (2023). Host interactions of novel Crassvirales species belonging to multiple families infecting bacterial host, Bacteroides cellulosilyticus WH2. Microbial Genomics, 9(9), 001100.
Park, G. W., Ng, T. F. F., Freeland, A. L., Marconi, V. C., Boom, J. A., Staat, M. A., & Vinjé, J. (2020). CrAssphage as a novel tool to detect human fecal contamination on environmental surfaces and hands. Emerging Infectious Diseases, 26(8), 1731.
Ramos-Barbero, M. D., Gómez-Gómez, C., Sala-Comorera, L., Rodríguez-Rubio, L., Morales-Cortes, S., Mendoza-Barberá, E., & Muniesa, M. (2023). Characterization of crAss-like phage isolates highlights Crassvirales genetic heterogeneity and worldwide distribution. Nature Communications, 14(1), 4295.
Rogier, R., Ederveen, T. H., Boekhorst, J., Wopereis, H., Scher, J. U., Manasson, J., & Abdollahi-Roodsaz, S. (2017). Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17-and TLR4-dependent arthritis. Microbiome, 5(1), 1–15.
Rossi, A., Treu, L., Toppo, S., Zschach, H., Campanaro, S., & Dutilh, B. E. (2020). Evolutionary study of the crassphage virus at gene level. Viruses, 12(9), 1035.
Sabar, M. A., Honda, R. & Haramoto, E. (2022). CrAssphage as an indicator of human-fecal contamination in water environment and virus reduction in wastewater treatment. Water Research, 221, 118827.
Sala-Comorera, L., Reynolds, L. J., Martin, N. A., Pascual-Benito, M., Stephens, J. H., Nolan, T. M., & Meijer, W. G. (2021). crAssphage as a human molecular marker to evaluate temporal and spatial variability in faecal contamination of urban marine bathing waters. Science of The Total Environment, 789, 147828.
Sangkaew, W., Kongprajug, A., Chyerochana, N., Ahmed, W., Rattanakul, S., Denpetkul, T., & Sirikanchana, K. (2021). Performance of viral and bacterial genetic markers for sewage pollution tracking in tropical Thailand. Water Research, 190, 116706.
Shahin, K., Soleimani-Delfan, A., He, Z., Sansonetti, P., & Collard, J. M. (2023). Metagenomics revealed a correlation of gut phageome with autism spectrum disorder. Gut Pathogens, 15(1), 1–7.
Shamash, M., & Maurice, C. F. (2022). Phages in the infant gut: A framework for virome development during early life. The ISME Journal, 16(2), 323–330.
Shkoporov, A. N., Khokhlova, E. V., Fitzgerald, C. B., Stockdale, S. R., Draper, L. A., Ross, R. P., & Hill, C. (2018). ΦCrAss001, a member of the most abundant bacteriophage family in the human gut, infects Bacteroides. bioRxiv. https://doi.org/10.1038/s41467-018-07225-7
Shkoporov, A. N., Khokhlova, E. V., Stephens, N., Hueston, C., Seymour, S., Hryckowian, A. J., & Hill, C. (2021). Long-term persistence of crAss-like phage crAss001 is associated with phase variation in Bacteroides intestinalis. BMC Biology, 19, 1–16.
Siranosian, B. A., Tamburini, F. B., Sherlock, G., & Bhatt, A. S. (2020). Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nature Communications, 11(1), 280.
Sresung, M., Paisantham, P., Ruksakul, P., Kongprajug, A., Chyerochana, N., Gallage, T. P., & Sirikanchana, K. (2023). Microbial source tracking using molecular and cultivable methods in a tropical mixed-use drinking water source to support water safety plans. Science of The Total Environment, 876, 162689.
Stachler, E., Akyon, B., de Carvalho, N. A., Ference, C., & Bibby, K. (2018). Correlation of crAssphage qPCR markers with culturable and molecular indicators of human fecal pollution in an impacted urban watershed. Environmental Science & Technology, 52(13), 7505–7512.
Stachler, E., & Bibby, K. (2014). Metagenomic evaluation of the highly abundant human gut bacteriophage CrAssphage for source tracking of human fecal pollution. Environmental Science & Technology Letters, 1(10), 405–409.
Stachler, E., Crank, K., & Bibby, K. (2019). Co-occurrence of crAssphage with antibiotic resistance genes in an impacted urban watershed. Environmental Science & Technology Letters, 6(4), 216–221.
Stachler, E., Kelty, C., Sivaganesan, M., Li, X., Bibby, K., & Shanks, O. C. (2017). Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environmental Science & Technology, 51(16), 9146–9154.
Stewart, L., Edgar, J. D. M., Blakely, G., & Patrick, S. (2018). Antigenic mimicry of ubiquitin by the gut bacterium Bacteroides fragilis: A potential link with autoimmune disease. Clinical & Experimental Immunology, 194(2), 153–165.
Tamburini, F. B., Sherlock, G. J., & Bhatt, A. S. (2018). Transmission and persistence of crAssphage, a ubiquitous human-associated bacteriophage. BioRxiv. https://doi.org/10.1101/460113v1
Tandukar, S., Sherchan, S. P., & Haramoto, E. (2020). Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Scientific Reports, 10(1), 3616.
Threndyle, R. E., Kurylyk, B. L., Huang, Y., Johnston, L. H., & Jamieson, R. C. (2022). CrAssphage as an indicator of groundwater-borne pollution in coastal ecosystems. Environmental Research Communications, 4(5), 051001.
Tomofuji, Y., Kishikawa, T., Maeda, Y., Ogawa, K., Otake-Kasamoto, Y., Kawabata, S., Nii, T., Okuno, T., Oguro-Igashira, E., Kinoshita, M., & Takagaki, M. (2022a). Prokaryotic and viral genomes recovered from 787 Japanese gut metagenomes revealed microbial features linked to diets, populations, and diseases. Cell Genomics, 2, 100219.
Tomofuji, Y., Kishikawa, T., Maeda, Y., Ogawa, K., Nii, T., Okuno, T., Oguro-Igashira, E., Kinoshita, M., Yamamoto, K., Sonehara, K., & Yagita, M. (2022b). Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Annals of the Rheumatic Diseases, 81(2), 278–288.
Turner, D., Shkoporov, A. N., Lood, C., Millard, A. D., Dutilh, B. E., Alfenas-Zerbini, P., & Adriaenssens, E. M. (2023). Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Archives of Virology, 168(2), 74.
Valério, E., Santos, M. L., Teixeira, P., Matias, R., Mendonça, J., Ahmed, W., & Brandão, J. (2022). Microbial source tracking as a method of determination of beach sand contamination. International Journal of Environmental Research and Public Health, 19(13), 7934.
Ward, L. M., Ghaju Shrestha, R., Tandukar, S., Sherchand, J. B., Haramoto, E., & Sherchan, S. P. (2020). Evaluation of crAssphage marker for tracking fecal contamination in river water in Nepal. Water, Air, & Soil Pollution, 231, 1–7.
Wilder, M. L., Middleton, F., Larsen, D. A., Du, Q., Fenty, A., Zeng, T., & Green, H. C. (2021). Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Research X, 11, 100100.
Wu, H., Juel, M. A. I., Eytcheson, S., Aw, T. G., Munir, M., & Molina, M. (2023). Temporal and spatial relationships of CrAssphage and enteric viral and bacterial pathogens in wastewater in North Carolina. Water Research, 239, 120008.
Wu, Z., Greaves, J., Arp, L., Stone, D., & Bibby, K. (2020). Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environment International, 136, 105452.
Yang, H., Gan, D., Li, Y., Wang, X., Jin, L., Qin, K., & Wang, Z. (2020). Quyushengxin formula causes differences in bacterial and phage composition in ulcerative colitis patients. Evidence-Based Complementary and Alternative Medicine. https://doi.org/10.1155/2020/585902
Yassour, M., Jason, E., Hogstrom, L. J., Arthur, T. D., Tripathi, S., Siljander, H., & Xavier, R. J. (2018). Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host & Microbe, 24(1), 146–154.
Yutin, N., Benler, S., Shmakov, S. A., Wolf, Y. I., Tolstoy, I., Rayko, M., & Koonin, E. V. (2021). Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nature Communications, 12(1), 1044.
Yutin, N., Makarova, K. S., Gussow, A. B., Krupovic, M., Segall, A., Edwards, R. A., & Koonin, E. V. (2018). Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nature Microbiology, 3(1), 38–46.
Zhang, Y., Wu, R., Li, W., Chen, Z., & Li, K. (2021). Occurrence and distributions of human-associated markers in an impacted urban watershed. Environmental Pollution, 275, 116654.
Zhang, Y., Wu, R., Lin, K., Wang, Y., & Lu, J. (2020). Performance of host-associated genetic markers for microbial source tracking in China. Water Research, 175, 115670.
Acknowledgements
The authors would like to express their sincere gratitude to the members of the manuscript review committee and the Director of ICMR—National Institute of Virology, Pune, Maharashtra, India, for their guidance and support. We want to acknowledge Ms. Meera Prabhakar, Bacteriology Group, ICMR—National Institute of Virology, Pune, Maharashtra, India, for helping to prepare the image included in the manuscript.
Funding
This review article received no external funding.
Author information
Authors and Affiliations
Contributions
Concept: RV, Review of literature: ATR and RV, first draft: ATR, Review and final draft: ATR and RV.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests related to the work submitted for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Remesh, A.T., Viswanathan, R. CrAss-Like Phages: From Discovery in Human Fecal Metagenome to Application as a Microbial Source Tracking Marker. Food Environ Virol (2024). https://doi.org/10.1007/s12560-024-09584-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12560-024-09584-5