Abstract
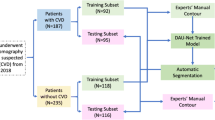
The aim of this study is to develop an automated deep-learning-based whole heart segmentation of ECG-gated computed tomography data. After 21 exclusions, CT acquired before transcatheter aortic valve implantation in 71 patients were reviewed and randomly split in a training (n = 55 patients), validation (n = 8 patients), and a test set (n = 8 patients). A fully automatic deep-learning method combining two convolutional neural networks performed segmentation of 10 cardiovascular structures, which was compared with the manually segmented reference by the Dice index. Correlations and agreement between myocardial volumes and mass were assessed. The algorithm demonstrated high accuracy (Dice score = 0.920; interquartile range: 0.906–0.925) and a low computing time (13.4 s, range 11.9–14.9). Correlations and agreement of volumes and mass were satisfactory for most structures. Six of ten structures were well segmented. Deep-learning-based method allowed automated WHS from ECG-gated CT data with a high accuracy. Challenges remain to improve right-sided structures segmentation and achieve daily clinical application.
Graphical abstract





Similar content being viewed by others
Abbreviations
- 3D:
-
Tridimensional
- WHS:
-
Whole heart segmentation
- CT:
-
Computed tomography
- AI:
-
Artificial intelligence
- TAVI:
-
Transcatheter aortic valve implantation
- PV:
-
Pulmonary veins
- LA:
-
Left atrium
- LVC:
-
Left ventricular cavity
- LVM:
-
Left ventricular myocardium
- Ao:
-
Aorta
- CS:
-
Coronary sinus
- SVC:
-
Superior vena cava
- RA:
-
Right atrium
- RVC:
-
Right ventricular cavity
- PA:
-
Pulmonary artery
- ROI:
-
Region of interest
- CNN:
-
Convolutional neural network
- IQR:
-
Interquartile
References
Siontis, G. C. M., Praz, F., Pilgrim, T., et al. (2016). Transcatheter aortic valve implantation vs. surgical aortic valve replacement for treatment of severe aortic stenosis: A meta-analysis of randomized trials. European Heart Journal, 37(47), 3503–3512. https://doi.org/10.1093/eurheartj/ehw225
Siontis, G. C. M., Overtchouk, P., Cahill, T. J., et al. (2019). Transcatheter aortic valve implantation vs. surgical aortic valve replacement for treatment of symptomatic severe aortic stenosis: An updated meta-analysis. European Heart Journal, 40(38), 3143–3153. https://doi.org/10.1093/eurheartj/ehz275
Auffret, V., Lefevre, T., Van Belle, E., et al. (2017). Temporal trends in transcatheter aortic valve replacement in France: FRANCE 2 to FRANCE TAVI. Journal of the American College of Cardiology, 70(1), 42–55. https://doi.org/10.1016/j.jacc.2017.04.053
Muller, D. W. M., Farivar, R. S., Jansz, P., et al. (2017). Transcatheter mitral valve replacement for patients with symptomatic mitral regurgitation: A global feasibility trial. Journal of the American College of Cardiology, 69(4), 381–391. https://doi.org/10.1016/j.jacc.2016.10.068 [published correction appears in J Am Coll Cardiol 2017;69(9):1213].
Guerrero, M., Urena, M., Himbert, D., et al. (2018). 1-Year outcomes of transcatheter mitral valve replacement in patients with severe mitral annular calcification. Journal of the American College of Cardiology, 71(17), 1841–1853. https://doi.org/10.1016/j.jacc.2018.02.054
Mas, J. L., Mas, J. L., Derumeaux, G., Guillon, B., et al. (2017). Patent foramen ovale closure or anticoagulation vs. antiplatelets after stroke. New England Journal of Medicine, 377(11), 1011–1021. https://doi.org/10.1056/NEJMoa1705915
Bishop, M., Rajani, R., Plank, G., et al. (2016). Three-dimensional atrial wall thickness maps to inform catheter ablation procedures for atrial fibrillation. Europace, 18(3), 376–383. https://doi.org/10.1093/europace/euv073
Blanke, P., Weir-McCall, J. R., Achenbach, S., et al. (2019). Computed tomography imaging in the context of transcatheter aortic valve implantation (TAVI)/transcatheter aortic valve replacement (TAVR): An expert consensus document of the Society of Cardiovascular Computed Tomography. JACC: Cardiovascular Imaging, 12(1), 1–24. https://doi.org/10.1016/j.jcmg.2018.12.003
Blanke, P., Dvir, D., Cheung, A., et al. (2015). Mitral annular evaluation with CT in the context of transcatheter mitral valve replacement. JACC: Cardiovascular Imaging, 8(5), 612–615. https://doi.org/10.1016/j.jcmg.2014.07.028
Blanke, P., Naoum, C., Dvir, D., et al. (2017). Predicting LVOT obstruction in transcatheter mitral valve implantation: Concept of the neo-LVOT. JACC: Cardiovascular Imaging, 10(4), 482–485. https://doi.org/10.1016/j.jcmg.2016.01.005
Weir-McCall, J. R., Blanke, P., Naoum, C., Delgado, V., Bax, J. J., & Leipsic, J. (2018). Mitral valve imaging with CT: Relationship with transcatheter mitral valve interventions. Radiology, 288(3), 638–655. https://doi.org/10.1148/radiol.2018172758
Zhuang, X., & Shen, J. (2016). Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Medical Image Analysis, 31, 77–87. https://doi.org/10.1016/j.media.2016.02.006
Commandeur, F., Goeller, M., Betancur, J., Cadet, S., Doris, M., Chen, X., et al. (2018). Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Transactions on Medical Imaging, 37, 1835–1846.
Zreik, M., Lessmann, N., van Hamersvelt, R. W., et al. (2018). Deep learning analysis of the myocardium in coronary CT angiography for identification of patients with functionally significant coronary artery stenosis. Medical Image Analysis, 44, 72–85. https://doi.org/10.1016/j.media.2017.11.008
Itu, L., Rapaka, S., Passerini, T., et al. (2016). A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. Journal of Applied Physiology1985, 121(1), 42–52. https://doi.org/10.1152/japplphysiol.00752.2015
Coenen, A., Kim, Y. H., Kruk, M., et al. (2018). Diagnostic accuracy of a machine-learning approach to coronary computed tomographic angiography-based fractional flow reserve: Result from the MACHINE consortium. Circulation: Cardiovascular Imaging, 11(6), e007217. https://doi.org/10.1161/CIRCIMAGING.117.007217
Zhuang, X., Bai, W., Song, J., et al. (2015). Multiatlas whole heart segmentation of CT data using conditional entropy for atlas ranking and selection. Medical Physics, 42(7), 3822–3833. https://doi.org/10.1118/1.4921366
Zhou, R., Liao, Z., Pan, T., et al. (2017). Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiotherapy and Oncology, 122(1), 66–71. https://doi.org/10.1016/j.radonc.2016.11.016
Cai, K., Yang, R., Chen, H., et al. (2017). A framework combining window width-level adjustment and Gaussian filter-based multi-resolution for automatic whole heart segmentation. Neurocomputing, 220, 138–150. https://doi.org/10.1016/j.neucom.2016.03.106
Zhuang, X., Li, L., Payer, C., et al. (2019). Evaluation of algorithms for multi-modality whole heart segmentation: An open-access grand challenge. Medical Image Analysis, 58, 101537. https://doi.org/10.1016/j.media.2019.101537
Baskaran, L., Maliakal, G., Al’Aref, S. J., et al. (2020). Identification and quantification of cardiovascular structures from CCTA: An end-to-end, rapid, pixel-wise, deep-learning method. JACC: Cardiovascular Imaging, 13(5), 1163–1171. https://doi.org/10.1016/j.jcmg.2019.08.025
Kaladji, A., Lucas, A., Kervio, G., Haigron, P., & Cardon, A. (2010). Sizing for endovascular aneurysm repair: Clinical evaluation of a new automated three-dimensional software. Annals of Vascular Surgery, 24(7), 912–920. https://doi.org/10.1016/j.avsg.2010.03.018
Lang, R. M., Badano, L. P., Mor-Avi, V., et al. (2015). Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. European Heart Journal – Cardiovascular Imaging, 16(3), 233–270. https://doi.org/10.1093/ehjci/jev014 [published correction appears in Eur Heart J Cardiovasc Imaging. 2016 Apr;17(4):412] [published correction appears in Eur Heart J Cardiovasc Imaging. 2016 Sep;17 (9):969].
Petersen, S. E., Khanji, M. Y., Plein, S., Lancellotti, P., & Bucciarelli-Ducci, C. (2019). European Association of Cardiovascular Imaging expert consensus paper: A comprehensive review of cardiovascular magnetic resonance normal values of cardiac chamber size and aortic root in adults and recommendations for grading severity. European Heart Journal - Cardiovascular Imaging, 20(12), 1321–1331. https://doi.org/10.1093/ehjci/jez232 [published correction appears in Eur Heart J Cardiovasc Imaging. 2019 Dec 1;20(12):1331].
Tobon-Gomez, C., Geers, A. J., Peters, J., et al. (2015). Benchmark for algorithms segmenting the left atrium from 3D CT and MRI datasets. IEEE Transactions on Medical Imaging, 34(7), 1460–1473. https://doi.org/10.1109/TMI.2015.2398818
Fuchs, A., Mejdahl, M. R., Kühl, J. T., et al. (2016). Normal values of left ventricular mass and cardiac chamber volumes assessed by 320-detector computed tomography angiography in the Copenhagen general population study. European Heart Journal Cardiovascular Imaging, 17(9), 1009–1017. https://doi.org/10.1093/ehjci/jev337
Forrest N. Landola, Song Han, Matthew W. Moskewicz, Khalid Ashraf, William J. Dally, Kurt Keutzer. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and < 0.50 MB model size. https://arxiv.org/abs/1602.07360. Accessed 18 April 2021
Karen Simonyan, Andrew Zisserman. Very deep convolutional network for large-scale image recognition. https://arxiv.org/abs/1409.1556. Accessed 18 April 2021
Gibson, E., Giganti, F., Hu, Y., et al. (2018). Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Transactions on Medical Imaging, 37(8), 1822–1834. https://doi.org/10.1109/TMI.2018.2806309
Eelbode, T., Bertels, J., Berman, M., et al. (2020). Optimization for medical image segmentation: Theory and practice when evaluating with dice score or Jaccard index. IEEE Transactions on Medical Imaging, 39(11), 3679–3690. https://doi.org/10.1109/TMI.2020.3002417
Chen, J., Yang, Z. G., Xu, H. Y., Shi, K., Long, Q. H., & Guo, Y. K. (2017). Assessments of pulmonary vein and left atrial anatomical variants in atrial fibrillation patients for catheter ablation with cardiac CT. European Radiology, 27(2), 660–670. https://doi.org/10.1007/s00330-016-4411-6
Cho J, Lee K, Shin E, Choy G, Do S. (2015) How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? http://arxiv.org/abs/151106348. Accessed 18 April 2021
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Animal Studies
No animal studies were carried out by the authors for this article.
Conflict of Interest
Dr. Vincent Auffret and Dr. Le Breton received lecture fees from Edwards Lifescience. Mr. Florent Lalys is one of the employees of Therenva®. The other authors have nothing to disclose. All the authors take responsibility for all aspects of the reliability and freedom from bias of the data presented and their discussed interpretation.
Additional information
Associate Editor Jozine ter Maaten oversaw the review of this article
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharobeem, S., Le Breton, H., Lalys, F. et al. Validation of a Whole Heart Segmentation from Computed Tomography Imaging Using a Deep-Learning Approach. J. of Cardiovasc. Trans. Res. 15, 427–437 (2022). https://doi.org/10.1007/s12265-021-10166-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-021-10166-0