Abstract
Epidemiological studies suggest that high fish intake is associated with a decreased risk of colorectal cancer which has been linked to the high content of the n-3 polyunsaturated fatty acids (PUFAs) eicosapentaenoic acids (EPA) and docosahexaenoic acid (DHA) in some fish. The aim of the study was to compare the modulation of gene expression in LT97 colon adenoma cells in response to EPA and DHA treatment. Therefore, we used custom-designed cDNA arrays containing probes for 306 genes related to stress response, apoptosis and carcinogenesis and hybridised them with cDNA from LT97 cells which were treated for 10 or 24 h with 50 μM EPA or DHA. There was a marked influence of n-3 PUFA on the expression of several gene types, such as detoxification, cell cycle control, signaling pathways, apoptosis and inflammation. DHA and EPA generally modulated different sets of genes, although a few common effects were noted. In our approach, we used preneoplastic adenoma cells which are a relevant model for target cells of chemoprevention. If verified with real time PCR, these results identify genes and targets for chemoprevention of colon cancer.
Similar content being viewed by others
Short communication
Epidemiological studies suggest that high fish intake is associated with a decreased risk of colorectal cancer (2, 3, 7). Most of the beneficial effects have been linked to the high content of the n-3 polyunsaturated fatty acids (PUFAs) eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) in some fish. EPA and DHA are essential for humans and found most notably in oily fish like salmon or herring. The intake of n-3 PUFAs in the world differs and is in direct relation to the relative fish consumption. The incidence of colorectal cancer in countries with high fish consumption such as Greenland or Japan is reported to be lower (9). Also, in vivo studies in rats have shown antitumourigenic effects of a diet rich in fish oil compared to other oils, such as corn oil (5). Evidence suggests that fish-related compounds act post-initiation to reduce transition of adenomas to tumours. However, the molecular mechanisms behind the anticancer effects of a diet rich in fish are not yet clarified.
The aim of the present study was to examine the modulation of expression of 306 genes related to stress response, apoptosis and carcinogenesis by comparing the effects of EPA and DHA. Human colon adenoma cells were chosen since they are a model for target cells for chemoprevention in vivo. Previous in vitro studies in this field have used cancer cell lines such as HT29 (1) or Caco-2 (6) rather than cells derived from adenomas, for example LT97.
Materials and methods
Gene expression studies were performed using LT97 human colon adenoma cells (4, 8) treated with a non-cytotoxic concentration (50 μM) of EPA (n = 3), DHA (n = 2) or alternatively 0.1% ethanol. The stock solution of each non-esterified fatty acid was purchased and dissolved in ethanol, thus ethanol was chosen to be the appropriate control. RNA was extracted after 10 and 24 h of incubation with the RNeasy Mini PLUS Kit (Qiagen GmbH, Hilden, Germany). RNA was quantified with the Nanodrop (Peqlab, Erlangen, Germany) and RNA integrity number (RIN) measured by Bioanalyzer (Agilent Technologies Deutschland GmbH, Böblingen, Germany). A control RNA which was labelled with Cy3 for each array was produced using untreated HT29 human colon carcinoma cells. Equal amounts of RNA (1 μg, RIN 8.5–10) were used for synthesising first strand of cDNA by Superscript II reverse transcriptase. A second strand cDNA was synthesized afterwards using Polymerase I, RNaseH and E.coli DNA ligase (Invitrogen GmbH, Karlsruhe, Germany). After clean-up of the product the double stranded cDNA was used for amplifying cRNA in order to increase the amount of RNA used for cDNA labelling with Cy3- and Cy5-dCTPs (GE Healthcare, Braunschweig, Germany). The reaction product was purified to yield clean, labelled and single stranded cDNA which was hybridised with a customised cDNA array (PIQOR, Miltenyi Biotec, Bergisch-Gladbach, Germany) containing 300 genes of interest and 6 housekeeping genes as described previously (11). Prior to this, the glass slide arrays were preheated and prehybridised. The final hybridisation step was carried out overnight in a water bath (65°C) in a humidified hybridisation chamber. After this, the custom arrays were washed and dried prior to scanning (MArS array reader, Ditabis, Pforzheim, Germany). Fluorescence intensities were analysed using the GeneSpotter software (MicroDiscovery, Berlin, Germany). LOESS-corrected raw data were normalised first in relation to the reference sample and second by setting the value for the housekeeping gene GAPDH equal to one. Data are presented as fold change (FC) compared to the respective ethanol control for each experiment.
Results and discussion
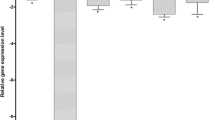
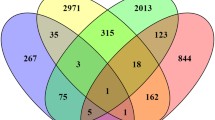
Using a stringent cut-off criteria (≤0.33, ≥2) for the obtained FC, 16 genes were differentially expressed in LT97 after treatment with DHA for 10 h (2 up, 14 down) and 14 (10 up, 4 down) after 24 h. Using the same criteria, treatment with EPA resulted in a higher number of differentially expressed genes after both treatment times: 83 (37 up, 46 down) genes were modulated after 10 h, 83 (78 up, 5 down) after 24 h. Examples of modulated genes are given in a summarised form in Table 1. Only a few genes were modulated by both DHA and EPA in the same manner: 9 of the genes were down-regulated by both PUFAs after 10 h and 2 of the genes were up-regulated after 24 h (see Fig. 1).
Venn diagram for cDNA results comparing effects of DHA (white circles) and EPA (grey circles) in LT97 after 10 (left panel) or 24 h (right panel) of treatment. The number of genes are shown, which are down- or up-regulated for indicated treatments, overlapping numbers are the result of genes which are regulated by both PUFA in the same manner
There was a marked influence of n-3 PUFA on the expression of several gene types (Table 1). Of these, some could result in functional effects such as altered biotransformation of phase I [e.g., up-regulation of CYP3A7 and FMO5 (24 h)] and phase II [up-regulation, e.g. GSTT2 (10), SULT1B1, CHST8, UGT1A family, UGT2B family down-regulation (10 h)], cell cycle control (e.g. up-regulation of cyclin dependent kinase 2), modulation of signalling pathways [e.g. down-regulation of Wnt3 (10 h), up-regulation of NFkB (24 h)], apoptosis [e.g., down-regulation of casp8 (10 h), bax up-regulation (24 h)], and inflammation (e.g. down-regulation of COX2). The modulation of gene expression by the n-3 PUFAs EPA and DHA has to be verified using real time PCR.
Our data show that n-3 PUFAs from fish oils may have a chemoprotective effect by modulating gene expression, which may prevent the development of adenomas to tumours. Our novel approach of studying this specific profile of gene expression modulation in preneoplastic human cells provides a rationale and relevant approach to identify target genes and agents that can contribute to chemoprevention.
References
Allred CD, Talbert DR et al (2008) PPARgamma1 as a molecular target of eicosapentaenoic acid in human colon cancer (HT-29) cells. J Nutr 138:250–256
Geelen A, Schouten JM et al (2007) Fish consumption, n-3 fatty acids, and colorectal cancer: a meta-analysis of prospective cohort studies. Am J Epidemiol 166:1116–1125
Hall MN, Chavarro JE et al (2008) A 22-year prospective study of fish, n-3 fatty acid intake, and colorectal cancer risk in men. Cancer Epidemiol Biomarkers Prev 17:1136–1143
Knoll N, Weise A et al (2006) 2-Dodecylcyclobutanone, a radiolytic product of palmitic acid, is genotoxic in primary human colon cells and in cells from preneoplastic lesions. Mutat Res 594:10–19
Latham P, Lund EK et al (1999) Dietary n-3 PUFA increases the apoptotic response to 1,2-dimethylhydrazine, reduces mitosis and suppresses the induction of carcinogenesis in the rat colon. Carcinogenesis 20:645–650
Narayanan BA, Narayanan NK et al (2001) Docosahexaenoic acid regulated genes and transcription factors inducing apoptosis in human colon cancer cells. Int J Oncol 19:1255–1262
Norat T, Bingham S et al (2005) Meat, fish, and colorectal cancer risk: the European prospective investigation into cancer and nutrition. J Natl Cancer Inst 97:906–916
Richter M, Jurek D et al (2002) Cells obtained from colorectal microadenomas mirror early premalignant growth patterns in vitro. Eur J Cancer 38:1937–1945
Swan J, Edwards BK (2003) Cancer rates among American Indians and Alaska natives: is there a national perspective. Cancer 98:1262–1272
Veeriah S, Balavenkatraman KK et al (2008a) Intervention with cloudy apple juice results in altered biological activities of ileostomy samples collected from individual volunteers. Eur J Nutr 47:226–234
Veeriah S, Miene C et al (2008b) Apple polyphenols modulate expression of selected genes related to toxicological defence and stress response in human colon adenoma cells. Int J Cancer 122:2647–2655
Acknowledgments
This paper is dedicated to the memory of Beatrice L. Pool-Zobel who passed away on 13 May 2008. Parts of this work have been funded by the Food Standard Agency’s project “BIOMICs”: (FSA N 12012). This work was performed within the Integrated Project SEAFOODplus, partially granted by the EU Commission under contract No FOOD-CT-2004-506359.
Conflict of interest statement
There are no authors’ conflict of interest which must be stated here.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Habermann, N., Lund, E.K., Pool-Zobel, B.L. et al. Modulation of gene expression in eicosapentaenoic acid and docosahexaenoic acid treated human colon adenoma cells. Genes Nutr 4, 73–76 (2009). https://doi.org/10.1007/s12263-009-0112-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12263-009-0112-y