Abstract
Polycomb proteins function in the maintenance of gene silencing via post-translational modifications of histones and chromatin compaction. Genetic and biochemical studies have revealed that the repressive function of Polycomb repressive complexes (PRCs) in transcription is counteracted by the activating function of Trithorax-group complexes; this balance fine-tunes the expression of genes critical for development and tissue homeostasis. The function of PRCs is frequently dysregulated in various cancer cells due to altered expression or recurrent somatic mutations in PRC genes. The tumor suppressive functions of EZH2-containing PRC2 and a PRC2-related protein ASXL1 have been investigated extensively in the pathogenesis of hematological malignancies, including myeloproliferative neoplasms (MPN). BCOR, a component of non-canonical PRC1, suppresses various hematological malignancies including MPN. In this review, we focus on recent findings on the role of PRCs in the pathogenesis of MPN and the therapeutic impact of targeting the pathological functions of PRCs in MPN.
Similar content being viewed by others
Introduction
Polycomb group (PcG) proteins were originally identified in Drosophila as regulators of body segmentation through the repression of homeotic genes and were subsequently identified in mammals. PcG proteins function in the maintenance of gene silencing via post-translational modifications of histones and chromatin compaction. In mammals, there are two major complexes of PcG complexes: Polycomb repressive complex 1 (PRC1) and 2 (PRC2), which modify mono-ubiquitination at lysine 119 of histone H2A (H2AK119ub1) and mono-, di-, and tri-methylation at lysine 27 of histone H3 (H3K27me1/me2/me3), respectively [1, 2]. Genetic and biochemical studies have revealed that the repressive function of PcG complexes in transcription is counteracted by the activating function of Trithorax-group (TrxG) complexes, which methylate H3K4. The functional balance between PcG and TrxG complexes fine-tunes expression of critical genes for development, adult tissue homeostasis [3], and stem cells [1]. On the other hand, dysregulated PRC results in a failure to maintain cellular homeostasis. The function of PRCs is frequently dysregulated in various cancer cells because of altered expression or recurrent somatic mutations in PRC genes, which have oncogenic and tumor suppressive roles that depend on cancer types [4, 5]. EZH2, which encodes the enzymatic component of PRC2, is one such PcG gene. The pathological role for deregulated EZH2 has been extensively investigated in various hematological malignancies, including myeloid malignancies, lymphoma, and acute T-cell lymphoblastic leukemia (T-ALL). Gain-of-function mutations in EZH2 play an oncogenic role in the development of B-cell lymphoma, while EZH2 also has a tumor suppressive role as missense and frame-shift mutations in EZH2 that abrogate its methyltransferase activity are frequently observed in MDS, MPN, and MDS/MPN overlap disorders [6]. There is also a tumor suppressive function of PRC1.1, a non-conical PRC1, in the pathogenesis of myeloid malignancies [7]. In this review, we focus on recent findings on the role of PRCs in the pathogenesis of myeloproliferative neoplasms (MPN) and the therapeutic impacts of targeting the pathological function of PRCs in MPN and related hematological malignancies.
Composition of Polycomb repressive complexes
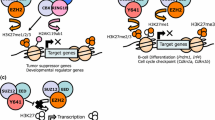
PRC2 contains three core subunits: SUZ12, EED, and one of the two methyltransferases, EZH1 or EZH2. PRC2 is recruited to non-methylated CpG islands (CGIs) and tri-methylates H3K27 (H3K27me3), which subsequently recruits canonical PRC1 via chromodomain proteins (CBX), a subunit of PRC1 that binds to H3K27me3 [8]. Canonical PRC1 contains four core subunits: PCGF4/BMI1 or PCGF2/MEL18, PHC, CBX and one of the two histone H2A mono-ubiquitylases, RING1A or RING1B, which has E3 ubiquitin ligase activity on histone H2A at lysine 119 (H2AK119ub1) thereby inducing the compaction of chromatin (Fig. 1) [9]. In addition to canonical PRC1, four non-canonical PRC1 variants (PRC1.1, PRC1.3, PRC1.5, and PRC1.6) containing a distinct PCGF subunit have been identified [10]. Non-canonical PRC1 complexes bind to target sites independently of PRC2-mediated H3K27me3 and deposit H2AK119ub1, which results in the subsequent recruitment of PRC2 and deposition of H3K27me3. In the case of PRC1.1, KDM2B binds to non-methylated CGIs through its DNA-binding domain and recruits other components of PRC1.1 (Fig. 1).
Canonical PcG complexes and non-canonical PRC1.1. PRC2 is recruited to non-methylated CGIs and tri-methylates H3K27 (H3K27me3), which subsequently recruits canonical PRC1 via chromodomain proteins (CBX), a subunit of PRC1 that binds to H3K27me3. Canonical PRC1 mono-ubiquitylates histone H2A at lysine 119 (H2AK119ub1), thereby inducing the compaction of chromatin (left panel). Non-canonical PRC1 complexes bind to target sites independently of PRC2-mediated H3K27me3 and deposit H2AK119ub1, which results in the subsequent recruitment of PRC2 and deposition of H3K27me3. In the case of PRC1.1, KDM2B binds to non-methylated CGIs through its DNA-binding domain and recruits other components of PRC1.1 (right panel)
Role of PRC in normal hematopoiesis
The biological function of canonical PRC1 and PRC2 has been characterized in detail in ES cells and hematopoietic stem cells (HSCs). Canonical PRC1 and PRC2 regulate the transcription of critical regulators for self-renewal and multipotency of HSCs to maintain homeostasis in hematopoiesis. The dysfunction of PRCs induced by targeted gene ablation of PRC component genes leads to the exhaustion of HSCs and/or impaired differentiation and production of blood cells [1, 11].
The loss of PRC2 function by the deletion of Eed markedly compromises adult hematopoiesis and impairs HSC function, in part, because of aberrant activation of Cdkn2a, a major target of PRCs that encodes p16Ink4a and p19Arf genes [12]. Ezh1 prevents premature senescence of HSCs via silencing the expression of Cdkn2a [13], while Ezh2 is dispensable for self-renewal of HSCs, at least in part, because of the compensatory function of Ezh1. Ezh1 co-regulates a large number of Ezh2 target genes and redistributes to a significant portion of Ezh2-specific targets in the context of Ezh2 insufficiency in normal and malignant hematopoietic stem cells [14, 15]. Ezh2-containing PRC2 (Ezh2-PRC2) also regulates the differentiation of B- and T-lymphoid cells and myeloid cells by regulating the expression of lineage-specific transcription factors [16]. These findings indicate that Ezh1-PRC2 and Ezh2-PRC2 coordinately regulate the transcription of PRC2 target genes but also have distinct functions in hematopoiesis.
PRC1.4, one of two canonical PRC1 that contain BMI1/PCGF4, is essential for the self-renewal capacity of HSCs via repressing expression of Cdkn2a [17]. Bmi1-deficient HSCs de-repress the expression of p16Ink4a and p19Arf. The deletion of both p16Ink4a and p19Arf largely rescues the self-renewal defect of Bmi1-deficient HSCs [18]. Conversely, overexpression of Bmi1 augments the function of HSCs by enhancing symmetrical cell division [18, 19]. PRC1.4 also restricts the ectopic expression of lineage-specific transcription factors, such as B cell master regulator genes Ebf1 and Pax5, in HSCs and multipotent progenitors (MPPs). Bmi1-deficient HSCs and MPPs have enhanced B cell commitment via premature activation of Ebf1 and Pax5 in HSCs and MPPs [20].
In contrast, the function of non-canonical PRC1 in hematopoiesis remains unclear. A genome-wide RNA interference screen found that Pcgf1, a component of PRC1.1, functions in concert with Runx1 transcription factor in Runx1-dependent differentiation. The depletion of both Runx1 and Pcgf1 resulted in the maintenance of proliferation of hematopoietic progenitor cells and perturbed their differentiation because of increased expression of posterior Hoxa cluster genes such as Hoxa9 [21]. Similarly, myeloid cells in mice lacking Bcor exons 9 and 10, which generates a truncated form of Bcor that does not bind Pcgf1, have a higher proliferative capacity, which results in myeloid-biased hematopoiesis [22, 23]. Bcor-deficient HSCs have increased expression of Cebp family genes in MPPs and sustained expression of posterior Hoxa cluster genes in myeloid progenitors [23]. These findings indicate that PCR1.1 negatively regulates critical transcriptional regulator genes, such as Cebp, and Hoxa9, to restrict the proliferation and differentiation of myeloid progenitor cells.
Additional sex combs like 1 (ASXL1) is a Polycomb-related protein frequently mutated in myeloid malignancies and clonal hematopoiesis in older healthy adults [24, 25]. ASXL1 regulates the function of Ezh2-PRC2 and modifies the function of BAP1, a nuclear deubiquitinase [26]. Hematopoietic-specific deletion of Asxl1 impairs the self-renewal capacity of HSCs, but results in multi-lineage cytopenia and dysplasia and increased numbers of stem and progenitor cells, which are characteristic features of MDS [27]. Loss of Asxl1 reduces global H3K27me3 levels and activates the expression of posterior Hoxa genes, such as Hoxa9, because of impaired recruitment of PRC2, which leads to the induction of MDS despite the reduced repopulating capacity of Asxl1-deficient HSCs [27,28,29] (Fig. 2).
ASXL1 mutant-induced epigenetic alterations. ASXL1 regulates the function of Ezh2-PRC2 and modifies the function of BAP1 in normal hematopoiesis. Loss of Asxl1 impairs recruitment of PRC2, which reduces H3K27me3 levels. In contrast, ASXL1 mutants enhance the de-ubiquitination activity of BAP1 on H2AK119, which significantly reduces H2AK119ub1 levels without affecting H3K27me3 levels. The ASXL1aa1–587 mutant interacts with BRD4, which increases the levels of H3K27ac and H3K122ac
Roles of PRC and PRC-related protein in myeloproliferative neoplasms
Classical MPNs include essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF). Driver mutations in JAK2, MPL, and CALR genes occur in all subtypes of MPNs. These mutations are generally considered mutually exclusive, but 10–15% of ET and PMF patients do not carry any of these mutations. In addition to these MPN driver gene mutations, it is important to identify mutation profiles of additional clonal marker genes, such as mutations in ASXL1, EZH2, TET2, IDH1/2, and spliceosome-related genes, because of their prognostic and therapeutic implications [30]. In addition, mutations in epigenetic modifiers promote the initiation and progression of MPN. We overview the pathogenic function of PRC genes including EZH2, ASXL1, and BCOR in the development of MPN.
Loss-of-function mutations in EZH2 promote the development of MPN
Although gain-of-function mutations in EZH2 have oncogenic roles in the development of B-cell lymphoma, missense and frame-shift mutations in EZH2 that abrogate its methyltransferase activity are frequently observed in MDS, MPN, and MDS/MPN overlap disorders (Table 1). These mutations reduce global levels of H3K27me3 and have been shown to increase expression levels of EZH2 target genes including potential oncogenes in tumor cells in patients and murine models, which indicate that EZH2 has a tumor suppressor function [4, 30, 31]. Loss-of-function mutations in EZH2 significantly predict poor outcomes in MDS, and the survival of patients with homozygous mutations is shorter than those with heterozygous mutations [32, 33]. Primary myelofibrosis (PMF) is a subtype of MPN driven by JAK2V617F activating mutations or CALR mutations that constitutively activate JAK2. EZH2 mutations independently predict poor survival in patients with PMF [34], consistent with the tumor suppressive function of EZH2. Monosomy 7 and deletion of the long arm of chromosome 7 (-7/7q-), characteristic cytogenetic anomalies frequently observed in myeloid malignancies, commonly involve EZH2 at 7q36 and are associated with the poor prognosis of PMF. A functional mapping study using 7q- MDS patient-derived iPS cells demonstrated that impaired production of hematopoietic cells is rescued by exogenous EZH2, which indicates that EZH2 haploinsufficiency contributes to the pathogenesis of 7q- MDS [35].
We and other groups have examined the impacts of loss-of-function mutations of EZH2 on the pathogenesis of myeloid malignancies using Ezh2 conditional knockout mice. Hematopoietic cell-specific deletion of Ezh2 resulted in the development of myeloid malignancies including MDS, MDS/MPN, and T-ALL, but not AML, after a long latency [14]. EZH2 mutations frequently occur with loss-of-function mutations in TET2 and RUNX1 in patients [36, 37]. Loss of Ezh2 enhances the formation of RUNX1 mutant-induced MDS, but inhibits leukemic transformation [38], which is consistent with an oncogenic role of Ezh2 observed in MLL-AF9-induced AML in which Ezh2 reinforces the transcriptional repression of myeloid-differentiation regulator genes [39, 40]. An Ezh2 deficiency (Ezh2Δ/Δ) in combination with a Tet2 hypomorph (Tet2KD/KD) in mice accelerates the transformation of HSCs and induces MDS and MDS/MPN [41]. Loss of Ezh2 significantly promotes the development of JAK2V617F mutant-induced myelofibrosis, at least in part, because of enhanced production of megakaryocytes with dysplastic features in BM [42,43,44]. These findings indicate that EZH2 fine-tunes the commitment and differentiation of HSCs, and EZH2 insufficiency promotes the transformation of HSCs to MDS and MPN stem cells.
Targeting therapy against EZH1 and EZH2 in myeloid malignancies
Because of the bimodal pathogenic functions of EZH2 as an oncogene and a tumor suppressor gene, pharmacological inhibition of EZH2 and both EZH1 and EZH2 has being tested extensively in pre-clinical and clinical studies in solid tumors, B-cell lymphoma, and AML [45]. An Ezh2 insufficiency aberrantly activates expression of certain oncogenes because of reduced levels of H3K27me3, but Ezh1-PRC2 partly compensates for Ezh2 loss in the maintenance of transcriptional repression of Ezh2 target genes [46]. In good agreement with these findings, AML cells are efficiently eradicated by the deletion of both Ezh1 and Ezh2 and treatment with EZH1/2 dual-inhibitors [47, 48], which indicates that Ezh1 is essential for the self-renewal capacity of leukemic stem cells in Ezh2-deficient conditions. These data support therapeutic approaches to target EZH1 and EZH2 using EZH1/2 dual inhibitors. A patient enrolled in a Phase I pediatric study (NCT02601937) of tazemetostat, an EZH2 inhibitor, for relapsed or refractory INI1 (a SWI/SNF component)-negative tumors or synovial sarcoma developed secondary T-cell lymphoblastic lymphoma (T-LBL), which suggests that caution is warranted when inhibiting the function of PRC2 for cancer therapy. However, the anti-tumor efficacies of Ezh2 and Ezh1/2 inhibitors are still encouraging [49]. Further investigation and comprehensive assessment of pre-clinical and clinical studies of Ezh2 and Ezh1/2 inhibitors are now underway.
Dysfunction of PRC2 sensitizes tumor cells to BRD4 inhibition
Because PRC2 represses target genes, loss-of-function mutations in EZH2 may de-repress expression of potential oncogenes in cancer. Indeed, loss of Ezh2 in JAK2V617F mice promotes an epigenetic switch characterized by reduced H3K27me3 levels followed by elevated H3K27 acetylation (H3K27ac) levels at promoter regions of PRC2 target genes, which results in the activation of potential oncogenes such as Hmga2 [42,43,44]. HMGA2 is significantly upregulated in CD34+ cells in PMF patients with EZH2 mutations [43, 50], and overexpression of Hmga2 in JAK2V617F HSCs increases the production of dysplastic megakaryocytes and the development of PMF in mice [42, 51]. Bromodomain inhibitors inhibit the function of enhancers by competitively interfering with the binding of BRD4 to H3K27ac and abrogates the progression of tumors [52]. Loss of PRC2 increases sensitivity to bromodomain inhibition of JAK2V617F myelofibrosis-initiating cells in vitro and in vivo [42]. Furthermore, a combination of bromodomain and JAK kinase inhibition reduces NF-kB-induced inflammation, which completely reverses fibrosis in JAK2V617F model mice [53]. Vulnerability to BRD4 inhibition has also been observed in solid tumors with compromised function of Ezh2-PRC2 by somatic gene mutations in PRC2 genes or in tumors expressing MLL1, which interacts with the p300/CBP complex to cause loss of H3K27me and gain of H3K27ac [54, 55]. Thus, these studies suggest that bromodomain inhibition in combination with JAK kinase inhibition is a novel therapeutic rationale for eradicating tumors and removing fibrosis in MPN patients.
Implication of DNA hypo-methylating therapy for MDS/MPN with EZH2 mutations
Promoter DNA hyper-methylation silences the expression of tumor suppressor genes, thereby promoting malignant transformation. Because MDS cells have higher levels of DNA hyper-methylation than de novo AML cells [56], hypomethylating agents, such as azacitidine (AZA) and decitabine (DAC), are clinically used to impede the progression of MDS in patients. However, the mutations of epigenetic modifiers and transcription factors, such as TET2 and TP53, do not always predict the response to AZA and DAC in patients with MDS, MPN, or AML [57,58,59]. PRC2 target genes defined in ES cells and normal HSPCs preferentially display DNA hyper-methylation in tumor cells, such as colon cancer, AML, and MDS, regardless of genetic mutations in epigenetic modifiers [60, 61]. Loss of Ezh2 significantly promotes the deposition of DNA methylation at promoter regions in many PRC2 target genes including development and differentiation regulator genes during the development of MDS and ETP-ALL in mice [46, 62]. Thus, PRC2 maintains a reversible gene silencing state of target genes by protecting them from aberrant DNA hyper-methylation. After mutation, deletion, or aberrant silencing of EZH2, alternative epigenetic machinery, such as promoter-hypermethylation, promotes transformation (Fig. 3).
EZH2 mutant-induced epigenetic alterations. PRC2 maintains a reversible gene silencing state of target genes by protecting them from aberrant DNA hyper-methylation. After mutation, deletion, or aberrant silencing of EZH2, alternative epigenetic machinery, such as promoter-hypermethylation, takes place and promotes transformation
Gain-of-function mutations in ASXL1 in the pathogenesis of myeloid malignancies
Somatic mutations in ASXL1 frequently mark hematological malignancies, such as AML, MDS, MPN, and CMML, and are associated with advanced age and poor clinical outcomes (Table 1) [32, 63]. Similar ASXL1 mutations are also found in clonal hematopoiesis in healthy older adults. Deletion of Asxl1 activates the expression of posterior Hoxa genes, such as Hoxa9, because of impaired recruitment of PRC2, which induces MDS despite the reduced repopulating capacity of Asxl1-deficient HSCs [27,28,29]. Nonsense or frameshift ASXL1 mutations generate truncated proteins that lack the C-terminus region [64] and are always heterozygous. These findings imply that ASXL1 mutations are gain-of-function or dominant-negative. BAP1, a deubiquitinase of H2AK119, assembles complexes with ASXL1, ASXL2, or ASXL3. BAP1 promotes mono-ubiquitination of ASXL1 mutants, which in turn enhance the deubiquitinase activity of BAP1 on H2AK119 and activates the expression of posterior Hoxa genes (Fig. 2) [65,66,67]. Expression of C-terminus-truncated ASXL1 mutants in mice induces myeloid malignancies, such as AML, MDS, MPN, and MDS/MPN, and promotes myeloid transformation in concert with additional mutations [68, 69]. In contrast to the reduced self-renewal capacity of Asxl1-deficient HSCs, HSCs with ASXLl1 mutants have sustained or enhanced competitive repopulating capacity in mice, and are accompanied by altered expression of genes critical for HSCs self-renewal and differentiation. Loss of Asxl1 reduces H3K27me3 levels but not H2AK119ub1 levels, while Asxl1 mutants significantly reduce H2AK119ub1 levels without affecting H3K27me3 levels (Fig. 2) [68]. Furthermore, a liquid chromatography-tandem mass spectrometry experiment revealed that an ASXL1aa1–587 mutant protein interacts with BRD4. Correspondingly, bone marrow hematopoietic cells expressing Asxl1Y588X have elevated levels of H3K27ac and H3K122ac and higher sensitivity to BRD4 inhibitors (Fig. 2) [69]. These findings suggest that ASXL1 mutants have a gain-of-function role in the pathogenesis of myeloid malignancies and provide a novel therapeutic strategy for ASXL1 mutant-induced malignancies.
Role of non-canonical PRC1.1 in the development of MPN
Among non-canonical PRC1 genes, loss-of-function mutations in BCOR occur in various hematological malignancies, such as AML, MDS, CMML, and lymphoid tumors (Table 1), indicating that BCOR functions as a tumor suppressor in these tumors [70,71,72] Mice deficient for Bcor exon 4 (Bcor∆E4/y), which generate a truncated protein that cannot bind Bcl6, develop Notch-dependent T-ALL [73]. Mice lacking Bcor exon 9 and exon 10 (Bcor∆E9−10/y), which cannot bind Pcgf1, develop T-ALL at a similar latency to that of Bcor∆E4/y mice and have myeloid-biased hematopoiesis. Bcor∆E9−10/y mice in combination with a Tet2 hypomorph develop lethal MDS, which transforms into MDS/MPN after a secondary transplantation. Bcor∆E9−10/y multipotent and myeloid progenitors have enhanced expression of Cebp family genes and posterior Hoxa cluster genes, respectively, because of reductions in H2AK119ub1 levels at the promoters of myeloid-regulator genes [23]. In addition, deletion of Kdm2b accelerates oncogenic KRAS-mediated myeloid transformation, and ectopic expression of Kdm2b suppresses the progression of KRAS-induced myeloid malignancies [74]. These findings suggest that PRC1.1 functions as a tumor suppressor in myeloid malignancies, such as MPN, in concert with other driver mutations. In contrast, several PRC1.1 component genes are overexpressed in AML cells in patients, and their knockdown significantly reduces the proliferative capacity of AML cells [75, 76], which suggests that PRC1.1 has opposing roles in tumor progression in a context-dependent manner.
Conclusion
PcG complexes have multiple epigenetic activities that differ depending on cell context, coexisting mutations, and tumor type. The impact of PcG complexes on transcription is complicated; they primarily function in transcriptional repression, but also activate transcription with non-PcG proteins. Therefore, it is important to determine how PRC dysfunction deregulates the expression of target genes and contributes to transformation. It is also important to understand alternative epigenetic pathways activated by PRC dysfunction such as epigenetic alteration of histone modifications and DNA methylation. A detailed understanding of PRC dysfunction will enable the development of new therapeutic approaches for individual cancer types by targeting novel therapeutic molecules or epigenetic pathways.
References
Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313.
Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16(11):643–9.
Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;293:1074–80.
Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612.
Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16(12):803–10.
Sashida G, Iwama A. Multifaceted role of the Polycomb-group gene EZH2 in hematological malignancies. Int J Hematol. 2017;105(1):23–30.
Isshiki Y, Iwama A. Emerging role of non-canonical Polycomb repressive complexes in normal and malignant hematopoiesis. Exp Hematol. 2018 (Published online: October 26, 2018).
Simon J, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–8.
Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, et al. PCGF Homologs, CBX Proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45(3):344–56.
Radulovic V, de Haan G, Klauke K. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia. 2013;27(3):523–33.
Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 2014;14(1):68–80.
Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11(5):649–62.
Mochizuki-Kashio M, Aoyama K, Sashida G, Oshima M, Tomioka T, Muto T, et al. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood. 2015;126(10):1172–83.
Aoyama K, Oshima M, Koide S, Suzuki E, Mochizuki-Kashio M, Kato Y, et al. Ezh1 targets bivalent genes to maintain self-renewing stem cells in Ezh2-insufficient myelodysplastic syndrome. iScience. 2018;9:161–74.
Su I, Dobenecker M-W, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121(3):425–36.
Park I, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–5.
Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203(10):2247–53.
Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the Polycomb gene product Bmi-1. Immunity. 2004;21(6):843–51.
Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the Polycomb protein Bmi1. Cell Stem Cell. 2010;6(3):279–86.
Ross K, Sedello AK, Todd GP, Paszkowski-rogacz M, Bird AW, Grinenko T, et al. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood. 2012;119(18):4152–62.
Cao Q, Gearhart MD, Gery S, Shojaee S, Yang H, Sun H, et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia. 2016;30(5):1155–65.
Tara S, Isshiki Y, Nakajima-Takagi Y, Oshima M, Aoyama K, Tanaka T, et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132(23):2470–83.
Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8.
Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose S, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87.
Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337(6101):1541–6.
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210(12):2641–59.
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through Loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–93.
Wang J, Li Z, He Y, Pan F, Chen S, Rhodes S, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123(4):541–53.
Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–79.
Iwama A. Polycomb repressive complexes in hematological malignancies. Blood. 2018;130(1):23–30.
Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–506.
Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–82.
Guglielmelli P, Biamonte F, Score J, Hidalgo-Curtis C, Cervantes F, Maffioli M, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118(19):5227–34.
Kotini AG, Chang C, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol. 2015;33(6):646–55.
Makishima H, Jankowska AM, Tiu RV, Szpurka H, Sugimoto Y, Hu Z, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24(10):1799–804.
Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia. 2013;27(6):1301–9.
Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5:4177.
Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120(5):1107–17.
Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci. 2012;109(13):5028–33.
Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210(12):2627–39.
Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, et al. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J Exp Med. 2016;213(8):1459–77.
Shimizu T, Kubovcakova L, Nienhold R, Zmajkovic J, Meyer SC, Shen HH, et al. Loss of Ezh2 synergizes with JAK2 -V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. J Exp Med. 2016;213(8):1479–96.
Yang Y, Akada H, Nath D, Hutchison RE, Mohi G. Loss of Ezh2 cooperates with Jak2V617F in the development of myelofibrosis in a mouse model of myeloproliferative neoplasm. Blood. 2016;127(26):3410–24.
Nakagawa M, Kitabayashi I. Oncogenic roles of enhancer of zeste homolog 1/2 in hematological malignancies. Cancer Sci. 2018;109(8):2342–8.
Hasegawa N, Oshima M, Sashida G, Matsui H, Koide S, Saraya A, et al. Impact of combinatorial dysfunctions of Tet2 and Ezh2 on the epigenome in the pathogenesis of myelodysplastic syndrome. Leukemia. 2017;31:861–71.
Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL- rearranged leukemia. Blood. 2015;125(2):346–57.
Fujita S, Honma D, Adachi N, Araki K, Takamatsu E, Katsumoto T, et al. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia. 2018;32(4):855–64.
Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19(5):649–59.
Guglielmelli P, Zini R, Bogani C, Salati S, Pancrazzi A, Bianchi E, et al. Molecular profiling of CD34 + cells in idiopathic myelofibrosis identifies a set of disease-associated genes and reveals the clinical significance of Wilms’ tumor gene 1 (WT1). Stem Cells. 2007;25(1):165–73.
Dutta A, Hutchison RE, Mohi G. Hmga2 promotes the development of myelofibrosis in Jak2(V617F) knockin mice by enhancing TGF-beta1 and Cxcl12 pathways. Blood. 2017;130(7):920–32.
Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–34.
Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, et al. Erratum: dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33(4):785–7.
De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514(7521):247–51.
Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X, et al. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-aberrant solid tumors. Cell. 2018;186–99.
Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448–58.
Bejar R, Lord A, Stevenson K, Bar-Natan M, Pérez-Ladaga A, Zaneveld J, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–12.
Meldi K, Qin T, Buchi F, Droin N, Sotzen J, Micol JB, et al. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest. 2015;125(5):1857–72.
Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375(21):2023–36.
Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39(2):237–42.
Gebhard C, Glatz D, Schwarzfischer L, Wimmer J, Stasik S, Nuetzel M, et al. Profiling of aberrant DNA methylation in acute myeloid leukemia reveals subclasses of CG-rich regions with epigenetic or genetic association. Leukemia. 2018;1–11.
Wang C, Oshima M, Sato D, Matsui H, Kubota S, Aoyama K, et al. Ezh2 loss propagates hypermethylation at T cell differentiation—regulating genes to promote leukemic transformation. J Clin Invest. 2018;128(9):3872–86.
Thol F, Friesen I, Damm F, Yun H, Weissinger EM, Krauter J, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29(18):2499–506.
Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, et al. Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations. J Clin Invest. 2013;123(11):4627–40.
Daou S, Barbour H, Ahmed O, Masclef L, Baril C, Sen Nkwe N, et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat Commun. 2018;9(1):4385.
Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:1–15.
Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, et al. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun. 2018;9(1):1–18.
Nagase R, Inoue D, Pastore A, Fujino T, Hou H-A, Yamasaki N, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215(6):1729–47.
Yang H, Kurtenbach S, Guo Y, Lohse I, Durante MA, Li J, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131(3):328–41.
Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118(23):6153–63.
Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y, et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122(18):3169–77.
Lasho TL, Mudireddy M, Finke CM, Hanson CA, Ketterling RP, Szuber N, et al. Targeted next-generation sequencing in blast phase myeloproliferative neoplasms. Blood Adv. 2018;2(4):370–80.
Tanaka T, Nakajima-Takagi Y, Aoyama K, Tara S, Oshima M, Saraya A, et al. Internal deletion of BCOR reveals a tumor suppressor function for BCOR in T lymphocyte malignancies. J Exp Med. 2017;214(10):2901–13.
Andricovich J, Kai Y, Peng W, Foudi A, Tzatsos A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J Clin Invest. 2016;126(3):905–20.
He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117(14):3869–80.
van den Boom V, Maat H, Geugien M, Rodríguez López A, Sotoca AM, Jaques J, et al. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep. 2016;14(2):332–46.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cell Fate” (#22118004) and “Stem Cell Aging and Disease” (#26115002) from MEXT, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Sashida, G., Oshima, M. & Iwama, A. Deregulated Polycomb functions in myeloproliferative neoplasms. Int J Hematol 110, 170–178 (2019). https://doi.org/10.1007/s12185-019-02600-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-019-02600-6