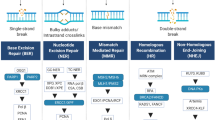
Abstract
Cancer-specific defects in DNA repair pathways create the opportunity to employ synthetic lethality approach. Recently, GEMA (gene expression and mutation analysis) approach detected insufficient expression of BRCA or NHEJ (non-homologous end joining) to predict PARP inhibitors response. We evaluated a possible role of DNA repair pathways using gene expression of single-strand break (XPA, XPC, XPG/ERCC5, CSA/ERCC8, and CSB/ERCC6) and double-strand break (ATM, BRCA1, BRCA2, RAD51, XRCC5, XRCC6, LIG4) in 92 patients with myelodysplastic syndrome (73 de novo, 9 therapy-related (t-MDS). Therapy-related MDS (t-MDS) demonstrated a significant downregulation of axis BRCA1-BRCA2-RAD51 comparing to normal controls (p = 0.048, p = 0.001, p = 0.001). XRCC6 showed significantly low expression in de novo MDS comparing to controls (p = 0.039) and for patients who presented chromosomal abnormalities (p = 0.047). Downregulation of LIG4 was consistently associated with poor prognostic markers in de novo MDS (hemoglobin < 8 g/dL (p = 0.040), neutrophils < 800/mm3 (p < 0.001), patients with excess of blasts (p = 0.001), very high (p = 0.002)/high IPSS-R (p = 0.043) and AML transformation (p < 0.001). We also performed an evaluation of GEPIA Database in 30 cancer types and detected a typical pattern of downregulation as here presented in primary or secondary MDS. All these results suggest synthetic lethality approach can be tested with DNA repair genes (beyond that of BRCA1/2 status) for de novo and therapy-related myelodysplastic syndrome and may encourage clinical trials evaluating the use of PARP1 inhibitors in MDS.
Graphic Abstract




Similar content being viewed by others
References
Schanz J, Cevik N, Fonatsch C, et al. Detailed analysis of clonal evolution and cytogenetic evolution patterns in patients with myelodysplastic syndromes (MDS) and related myeloid disorders. Blood Cancer J. 2018;8(3):28. https://doi.org/10.1038/s41408-018-0061-z.
Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. https://doi.org/10.1182/blood-2016-03-643544.
Chua CC, Fleming S, Wei AH. Clinicopathological aspects of therapy-related acute myeloid leukemia and myelodysplastic syndrome. Best Pract Res Clin Haematol. 2019;32(1):3–12. https://doi.org/10.1016/j.beha.2019.02.007.
Vargas-Rondón N, Villegas VE, Rondón-Lagos M. The role of chromosomal instability in cancer and therapeutic responses. Cancers (Basel). 2017. https://doi.org/10.3390/cancers10010004.
Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–85. https://doi.org/10.1056/NEJMra0804615.
Greenberg PL. Synergistic interactions of molecular and clinical advances for characterizing the myelodysplastic syndrome. J Natl Compr Cancer Netw. 2015;13:829–32.
Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355(6331):1330–4. https://doi.org/10.1126/science.aaf9011.
Ribeiro HL Jr, de Oliveira RT, Maia AR, et al. Polymorphisms of DNA repair genes are related to the pathogenesis of myelodysplastic syndrome. Hematol Oncol. 2015;33:220–8. https://doi.org/10.1002/hon.2175.
Valka J, Vesela J, Votavova H, et al. Differential expression of homologous recombination DNA repair genes in the early and advanced stages of myelodysplastic syndrome. Eur J Haematol. 2017;99(4):323–31. https://doi.org/10.1111/ejh.12920.
Scharer OS. Nucleotide excision repair in eukaryotes. Department of Pharmacological Sciences and Department of Chemistry, Stony Brook University, Stony Brook, New York 11974-3400. Cold Spring Harb Perspect Biol. 2013;5:a012609. https://doi.org/10.1101/cshperspect.a012609.
Alekseev S, Coin F. Orchestral maneuvers at the damaged sites in nucleotide excision repair. Cell Mol Life Sci. 2015;72(11):2177–86. https://doi.org/10.1007/s00018-015-1859-5.
Wouter LL, Nicolaas GJJ, Hoeijmakers JHJ. Molecular mechanism of nucleotide excision repair. Genes Dev. 2016. https://doi.org/10.1101/gad.13.7.768.
Saijo M. The role of Cockayne syndrome group A (CSA) protein in transcription-coupled nucleotide excision repair. Mech Ageing Dev. 2013;134:196–201. https://doi.org/10.1016/j.mad.2013.03.008.
Petruseva IO, Evdokimov AN, Lavrik OI. Molecular mechanism of global genome nucleotide excision repair. Acta Nat. 2014;6(1):20.
Mandal PK, Blanpain C, Rossi DJ. DNA damage response in adult stem cells: pathways and consequences. Nat Rev Mol Cell Biol. 2011;12:198–202. https://doi.org/10.1038/nrm3060.
Meht AA, Habe RJE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014. https://doi.org/10.1101/cshperspect.a016428.
Bhargava R, Onyango DO, Stark JM. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016;32(9):566–75. https://doi.org/10.1016/j.tig.2016.06.007.
Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7(6):7–152. https://doi.org/10.1186/1471-2407-7-152.
Economopoulou P, Pappa V, Kontsioti F, et al. Expression analysis of proteins involved in the non homologous end joining DNA repair mechanism in the bone marrow of adult de novo myelodysplastic syndromes. Ann Hematol. 2010;89(3):233–9. https://doi.org/10.1007/s00277-009-0823-6.
Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. https://doi.org/10.1038/nature03445.
Pinheiro RF, Chauffaille ML. Comparison of I-FISH and G-banding for the detection of chromosomal abnormalities during the evolution of myelodysplastic syndrome. Braz J MedBiol Res. 2009;42(11):1110–2. https://doi.org/10.1590/S0100-879X2009001100018.
McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: an international system for human cytogenomic nomenclature (2016). Cytogenet Genom Res. 2016;149:1–2.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(−Delta DeltaC(T)) method. Methods. 2001;25:402–8. https://doi.org/10.1006/meth.2001.1262.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8.
Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017. https://doi.org/10.1093/nar/gkx247.10.1093/nar/gkx247.
Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–39. https://doi.org/10.1016/j.tig.2014.06.003.
Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72(11):2814–21. https://doi.org/10.1158/0008-5472.CAN-11-3417.
Metzger MJ, Stoddard BL, Monnat RJ. PARP mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks. DNA Repair. 2013;12(7):529–34. https://doi.org/10.1016/j.dnarep.2013.04.004.
Nieborowska-Skorska M, Sullivan K, Dasgupta Y, et al. Gene expression and mutation-guided synthetic lethality eradicates proliferating and quiescent leukemia cells. J Clin Invest. 2017;127(6):2392–406. https://doi.org/10.1172/JCI90825.
Faraoni I, Graziani G. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers (Basel). 2018;10(12):487. https://doi.org/10.3390/cancers10120487.
Funding
This study was partially supported by the National Council of Technological and Scientific Development (CNPq) (Grant Nos. #420501/2018-5 and #424542/2016-1).
Author information
Authors and Affiliations
Contributions
HLRJ, RTGO, SMMM and RFP designed the study, provided patient materials and were responsible for collection and assembly of data. HLRJ, RTGO, AWAS, MBC, IRF and DPB performed the molecular procedures and analyzed the data. All drafted and edited the manuscript. All authors have approved the final version of manuscript before publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethical approval
All procedures were approved by the Ethics Committee of UFC (#1.292.509) and are in accordance with the 1964 Helsinki declaration and its later amendments.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Junior, H.L.R., de Oliveira, R.T.G., de Paula Borges, D. et al. Can synthetic lethality approach be used with DNA repair genes for primary and secondary MDS?. Med Oncol 36, 99 (2019). https://doi.org/10.1007/s12032-019-1324-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-019-1324-7