Abstract
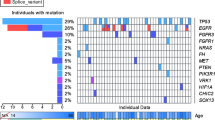
One hundred and ninety-five consecutive surgically treated Russian colorectal cancer (CRC) patients were retrospectively analyzed for the presence of mutations in KRAS, NRAS, BRAF and PIK3CA genes as well as for the microsatellite instability status. Comparison between high-resolution melting analysis, co-amplification at lower denaturation temperature PCR, DNA sequencing and allele-specific PCR for the detection of KRAS codon 12/13 mutations revealed that none of these methods alone provided satisfactory results in 100 % of the analyzed cases; this experience supports the use of more than one mutation-detecting technique at least in some circumstances. KRAS codon 12/13 substitutions were detected in 70 (35.9 %) CRC cases. Other mutations in the RAS/RAF genes occurred in 22 (11.3 %) cases and included rare KRAS (n = 6), NRAS (n = 8) and BRAF (n = 8) alterations. 5 BRAF mutations affected codon 600, while the remaining 3 potentially functional substitutions were located in the position 594. Twenty-four (12.3 %) CRC cases carried mutations in the PIK3CA, and 18 of these tumors also contained activating alteration in the RAS/RAF genes (p = 0.007). Only 3 (1.5 %) CRC cases showed high-level microsatellite instability (MSI-H) as determined by a panel of mononucleotide markers. Overall, the distribution of potentially predictive mutations in Russian CRC cases is similar to the one observed in other patient series of European descent. Noticeable occurrence of D594G mutation in BRAF oncogene and low frequency of MSI-H may deserve specific attention.

Similar content being viewed by others
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C and Parkin DM.GLOBOCAN 2008 v2.0. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet]. Lyon, France: International Agency for Research on Cancer. 2010. Available from http://globocan.iarc.fr.
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–47.
De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603.
Berg M, Soreide K. EGFR and downstream genetic alterations in KRAS/BRAF and PI3K/AKT pathways in colorectal cancer: implications for targeted therapy. Discov Med. 2012;14:207–14.
Ren J, Li G, Ge J, Li X, Zhao Y. Is K-ras gene mutation a prognostic factor for colorectal cancer: a systematic review and meta-analysis. Dis Colon Rectum. 2012;55:913–23.
Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. PLoS One. 2012. doi: 10.1371/journal.pone.0047054.
Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5.
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34.
Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–9.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62.
Lin JS, Webber EM, Senger CA, Holmes RS, Whitlock EP. Systematic review of pharmacogenetic testing for predicting clinical benefit to anti-EGFR therapy in metastatic colorectal cancer. Am J Cancer Res. 2011;1:650–62.
Mao C, Liao RY, Qiu LX, Wang XW, Ding H, Chen Q. BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis. Mol Biol Rep. 2011;38:2219–23.
Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group, Calonge N, Fisher NL, Berg AO, Campos-Outcalt D, Djulbegovic B, Ganiats TG, Haddow JE, Klein RD, Lyman DO, Offit K, Pauker SG, Piper M, Richards CS, Strickland OL, Tunis SR, Veenstra DL. Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy? Genet Med. 2013;. doi:10.1038/gim.2012.184.
Peeters M, Oliner KS, Parker A, Siena S, Van Cutsem E, Huang J, Humblet Y, Van Laethem JL, André T, Wiezorek J, Reese D, Patterson SD. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res. 2013;19:1902–12.
Wu S, Gan Y, Wang X, Liu J, Li M, Tang Y. PIK3CA mutation is associated with poor survival among patients with metastatic colorectal cancer following anti-EGFR monoclonal antibody therapy: a meta-analysis. J Cancer Res Clin Oncol. 2013;139:891–900.
Xu Q, Xu AT, Zhu MM, Tong JL, Xu XT, Ran ZH. Predictive and prognostic role of BRAF mutation in metastatic colorectal cancer patients treated with anti-epidermal growth factor receptor monoclonal antibodies: a meta-analysis. J Dig Dis. 2013;. doi:10.1111/1751-2980.12063.
Soliman AS, Bondy ML, El-Badawy SA, Mokhtar N, Eissa S, Bayoumy S, Seifeldin IA, Houlihan PS, Lukish JR, Watanabe T, Chan AO, Zhu D, Amos CI, Levin B, Hamilton SR. Contrasting molecular pathology of colorectal carcinoma in Egyptian and Western patients. Br J Cancer. 2001;85:1037–46.
Kislitsin D, Lerner A, Rennert G, Lev Z. K-ras mutations in sporadic colorectal tumors in Israel: unusual high frequency of codon 13 mutations and evidence for nonhomogeneous representation of mutation subtypes. Dig Dis Sci. 2002;47:1073–9.
Boardman LA, Lanier AP, French AJ, Schowalter KV, Burgart LJ, Koller KR, McDonnell SK, Schaid DJ, Thibodeau SN. Frequency of defective DNA mismatch repair in colorectal cancer among the Alaska Native people. Cancer Epidemiol Biomarkers Prev. 2007;16:2344–50.
Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A, Raeburn S, Lee E, Giardiello F, Smoot DT, Vilkin A, Boland CR, Goel A, Hafezi M, Nouraie M, Ashktorab H. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68.
Palomba G, Colombino M, Contu A, Massidda B, Baldino G, Pazzola A, Ionta M, Capelli F, Trova V, Sedda T, Sanna G, Tanda F, Budroni M, Sardinian Translational Oncology Group (STOG), Palmieri G, Cossu A, Contu M, Cuccu A, Farris A, Macciò A, Mameli G, Olmeo N, Ortu S, Petretto E, Pusceddu V, Virdis L. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med. 2012;10:178.
Smith G, Bounds R, Wolf H, Steele RJ, Carey FA, Wolf CR. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours—implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703.
André T, Blons H, Mabro M, Chibaudel B, Bachet JB, Tournigand C, Bennamoun M, Artru P, Nguyen S, Ebenezer C, Aissat N, Cayre A, Penault-Llorca F, Laurent-Puig P, de Gramont A, GERCOR. Panitumumab combined with irinotecan for patients with KRAS wild-type metastatic colorectal cancer refractory to standard chemotherapy: a GERCOR efficacy, tolerance, and translational molecular study. Ann Oncol. 2013;24:412–9.
Tol J, Dijkstra JR, Vink-Börger ME, Nagtegaal ID, Punt CJ, Van Krieken JH, Ligtenberg MJ. High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med. 2010;14:2122–31.
Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–8.
Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23:1518–25.
Macedo MP, de Andrade LB, Coudry R, Crespo R, Gomes M, Lisboa BC, Aguiar S Jr, Soares FA, Carraro DM, Cunha IW. Multiple mutations in the Kras gene in colorectal cancer: review of the literature with two case reports. Int J Colorectal Dis. 2011;26:1241–8.
Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun R, Nosho K, Meyerhardt JA, Giovannucci E, Fuchs CS, Chan AT, Ogino S. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606.
Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87.
Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–62.
Perucho M. Correspondence re: C.R. Boland et al., A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257,. Cancer Res. 1999;59:249–256.
Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene. 2008;27:6313–21.
Acknowledgments
This work has been supported by the Russian Foundation for Basic Research (Grants 11-04-01643, 12-04-00535, 12-04-00928 and 12-04-31576), Russian Ministry of Education and Science (contract 14.512.11.0041), the Dynasty Foundation (contract 18/13), the President’s Research Council for Support of Young Russian Scientists (fellowships 768.2012.4 and 790.2012.4) and the Grant from Amgen Inc. (Thousand Oaks, CA).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yanus, G.A., Belyaeva, A.V., Ivantsov, A.O. et al. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol 30, 686 (2013). https://doi.org/10.1007/s12032-013-0686-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-013-0686-5