Abstract
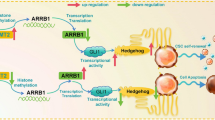
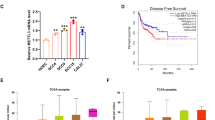
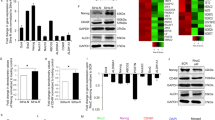
The major problem to effective treatment of oral cancer is the presence of therapy resistance. Presence of cancer stem cell in the bulk of tumor have been implicated in therapeutic resistance. In this study, we report a non-telomeric role of TRF2 in formation of oral cancer spheroids and CSC phenotype maintenance via an efficient DNA damage repair mechanism in the presence of chemotherapeutic insult. We report reduced sphere formation efficiency and reduced spheroid size in TRF2 silenced oral cancer cell lines. TRF2 silenced orospheres further reported reduced proliferative capacity as compared to non-silenced orospheres. Furthermore, TRF2 silencing hampered the migratory potential of oral cancer cell line and also reduced the expression of several CSC markers like CD44, Oct4, Sox2, KLF4 and c-Myc along with β-catenin and hTERT molecules both in Cal27 cell line and generated orospheres. TRF2 silencing impaired efficient DNA damage repair capacity of non-orospheric and orospheric cells and repressed ERCC1 expression levels when treated with Cisplatin. TRF2 overexpression was also observed to correlate with poor overall survival and disease relapse of OSCC patients. In silico studies further identified several amino acid residues that show high binding affinity and strong protein-protein interactions among TRF2 and CSC marker KLF4. Hence, our report confirms a non-telomeric role of TRF2 in spheroid generation, maintenance of CSC phenotype and efficient DNA damage repair capacity contributing to chemotherapy resistance in oral cancer cell line. We further iterate the use of TRF2 as a prognostic marker in OSCC for faster detection and improved survival.








Similar content being viewed by others
References
Padhi, S., Saha, A., Kar, M., Ghosh, C., Adhya, A., Baisakh, M., Mohapatra, N., Venkatesan, S., Hande, M. P., & Banerjee, B. (2015). Clinico-pathological correlation of β-catenin and telomere dysfunction in head and neck squamous cell carcinoma patients. Journal of Cancer, 6, 192–202.
Kekatpure, V. K. M. (2010). Oral Cancer in India: Learning from different populations. Cancer Prevention.
González-Moles, M. A., Scully, C., Ruiz-Ávila, I., & Plaza-Campillo, J. J. (2013). The cancer stem cell hypothesis applied to oral carcinoma. Oral Oncology, 49, 738–746.
Krishnamurthy, S., & Head, N. J. E. (2012). Neck Cancer stem cells. Journal of Dental Research, 91, 334–340.
Chinn, S. B., Darr, O. A., Owen, J. H., Bellile, E., McHugh, J. B., Spector, M. E., Papagerakis, S. M., Chepeha, D. B., Bradford, C. R., Carey, T. E., & Prince, M. E. P. (2015). Cancer stem cells: Mediators of tumorigenesis and metastasis in head and neck squamous cell carcinoma. Head & Neck, 37, 317–326.
Naik, P. P., Das, D. N., Panda, P. K., Mukhopadhyay, S., Sinha, N., Praharaj, P. P., Agarwal, R., & Bhutia, S. K. (2016). Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncology, 62, 122–135.
Sukach, A. N., & Ivanov, E. N. (2007). Formation of spherical colonies as a property of stem cells. Cell and Tissue Biology, 1, 476–481.
Fujii, H., Honoki, K., Tsujiuchi, T., Kido, A., Yoshitani, K., & Takakura, Y. (2009). Sphere-forming stem-like cell populations with drug resistance in human sarcoma cell lines. International Journal of Oncology, 34, 1381–1386.
Qiu, X., Wang, Z., Li, Y., Miao, Y., Ren, Y., & Luan, Y. (2012). Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. Cancer Letters, 323, 161–170.
Castillo, V., Valenzuela, R., Huidobro, C., Contreras, H. R., & Castellon, E. A. (2014). Functional characteristics of cancer stem cells and their role in drug resistance of prostate cancer. International Journal of Oncology, 45, 985–994.
Iwai, S., Yonekawa, A., Harada, C., Hamada, M., Katagiri, W., Nakazawa, M., & Yura, Y. (2010). Involvement of the Wnt-beta-catenin pathway in invasion and migration of oral squamous carcinoma cells. International Journal of Oncology, 37, 1095–1103.
Prosperi, J. R., Luu, H. H., & Goss, K. H. (2011). Dysregulation of the Wnt pathway in solid tumors. In K. H. Goss & M. Kahn (Eds.), Targeting the Wnt pathway in Cancer (pp. 81–128). New York, NY: Springer New York.
Lee, S. H., Koo, B. S., Kim, J. M., Huang, S., Rho, Y. S., Bae, W. J., Kang, H. J., Kim, Y. S., Moon, J. H., & Lim, Y. C. (2014). Wnt/β-catenin signalling maintains self-renewal and tumourigenicity of head and neck squamous cell carcinoma stem-like cells by activating Oct4. The Journal of Pathology, 234, 99–107.
Chen, Y., Shi, L., Zhang, L., Li, R., Liang, J., Yu, W., Sun, L., Yang, X., Wang, Y., Zhang, Y., & Shang, Y. (2008). The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. The Journal of Biological Chemistry, 283, 17969–17978.
Cai, C., & Zhu, X. (2012). The Wnt/beta-catenin pathway regulates self-renewal of cancer stem-like cells in human gastric cancer. Molecular Medicine Reports, 5, 1191–1196.
Diala, I., Wagner, N., Magdinier, F., Shkreli, M., Sirakov, M., Bauwens, S., Schluth-Bolard, C., Simonet, T., Renault, V. M., Ye, J., Djerbi, A., Pineau, P., Choi, J., Artandi, S., Dejean, A., Plateroti, M., & Gilson, E. (2013). Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Reports, 14, 356–363.
Hoffmeyer, K., Raggioli, A., Rudloff, S., Anton, R., Hierholzer, A., del Valle, I., Hein, K., Vogt, R., & Kemler, R. (2012). Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science (New York, N.Y.), 336, 1549–1554.
Feuerhahn, S., Chen, L. Y., Luke, B., & Porro, A. (2015). No DDRama at chromosome ends: TRF2 takes Centre stage. Trends in Biochemical Sciences, 40, 275–285.
Tanaka, H., Mendonca, M. S., Bradshaw, P. S., et al. (2005). DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proceedings of the National Academy of Sciences of the United States of America, 15539–15544.
Bradshaw, P. S., Stavropoulos, D. J., & Meyn, M. S. (2005). Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nature Genetics, 37, 193–197.
Mao, Z., Seluanov, A., Jiang, Y., & Gorbunova, V. (2007). TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America, 104, 13068–13073.
Saha, A. P. S., Kar, M., Roy, S., Maiti, P., & Banerjee, B. (2017). Role of TRF2 in efficient DNA repair, spheroid formation and Cancer stem cell maintenance. Oncomedicine, 2, 71–79.
Anaya, J. (2016). OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer Journal of Computer Science, 2, e67.
Borowicz, S., Van Scoyk, M., Avasarala, S., et al. (2014). The Soft Agar Colony Formation Assay., e51998.
Saha, A., Shree Padhi, S., Roy, S., & Banerjee, B. (2014). Method of detecting new cancer stem cell-like enrichment in development front assay (DFA). Journal of Stem Cells, 9, 235–242.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408.
Comeau, S. R., Gatchell, D. W., Vajda, S., & Camacho, C. J. (2004). ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Research, 32, W96–W99.
Wallace, A. C., Laskowski, R. A., & Thornton, J. M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8, 127–134.
Jenik, M., Parra, R. G., Radusky, L. G., Turjanski, A., Wolynes, P. G., & Ferreiro, D. U. (2012). Protein frustratometer: A tool to localize energetic frustration in protein molecules. Nucleic Acids Research, 40, W348–W351.
Siddik, Z. H. (2003). Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene, 22, 7265–7279.
Zamble, D. B., & Lippard, S. J. (1995). Cisplatin and DNA repair in cancer chemotherapy. Trends in Biochemical Sciences, 20, 435–439.
Niedernhofer, L. J., Odijk, H., Budzowska, M., van Drunen, E., Maas, A., Theil, A. F., de Wit, J., Jaspers, N. G. J., Beverloo, H. B., Hoeijmakers, J. H. J., & Kanaar, R. (2004). The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Molecular and Cellular Biology, 24, 5776–5787.
Britten, R. A., Liu, D., Tessier, A., Hutchison, M. J., & Murray, D. (2000). ERCC1 expression as a molecular marker of cisplatin resistance in human cervical tumor cells. International Journal of Cancer, 89, 453–457.
Arora, S., Kothandapani, A., Tillison, K., Kalman-Maltese, V., & Patrick, S. M. (2010). Downregulation of XPF–ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair, 9, 745–753.
McNeil, E. M., & Melton, D. W. (2012). DNA repair endonuclease ERCC1–XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Research, 40, 9990–10004.
Zhu, X. D., Niedernhofer, L., Kuster, B., Mann, M., Hoeijmakers, J. H., & de Lange, T. (2003). ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Molecular Cell, 12, 1489–1498.
Munoz, P., Blanco, R., Flores, J. M., & Blasco, M. A. (2005). XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nature Genetics, 37, 1063–1071.
Wu, Y., Mitchell, T. R. H., & Zhu, X.-D. (2008). Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mechanisms of Ageing and Development, 129, 602–610.
Liu, X., Huang, J., Chen, T., Wang, Y., Xin, S., Li, J., Pei, G., & Kang, J. (2008). Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Research, 18, 1177–1189.
Müller, M., Hermann, P. C., Liebau, S., Weidgang, C., Seufferlein, T., Kleger, A., & Perkhofer, L. (2016). The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Research, 16, 349–357.
Koo, B. S., Lee, S. H., Kim, J. M., Huang, S., Kim, S. H., Rho, Y. S., Bae, W. J., Kang, H. J., Kim, Y. S., Moon, J. H., & Lim, Y. C. (2015). Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene, 34, 2317–2324.
Yu, F., Li, J., Chen, H., Fu, J., Ray, S., Huang, S., Zheng, H., & Ai, W. (2011). Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene, 30, 2161–2172.
Gabay, M., Li, Y., & Felsher, D. W. (2014). MYC activation is a Hallmark of Cancer initiation and maintenance. Cold Spring Harbor Perspectives in Medicine, 4, a014241.
Ghuwalewala, S., Ghatak, D., Das, P., Dey, S., Sarkar, S., Alam, N., Panda, C. K., & Roychoudhury, S. (2016). CD44highCD24low molecular signature determines the Cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Research, 16, 405–417.
Fujiwara, Y., Yoshikawa, R., Tsujie, M., et al. (2015). CD44, a cancer stemness marker, to predict poor clinical outcome of esophageal cancer patients after neoadjuvant chemoradiotherapy. Journal of Clinical Oncology, 33, e15068-e.
Chanmee, T., Ontong, P., Kimata, K., & Itano, N. (2015). Key roles of Hyaluronan and its CD44 receptor in the Stemness and survival of Cancer stem cells. Frontiers in Oncology, 5, 180.
Shi Y, Ai W. (2013) Function of KLF4 in stem cell biology. In: Bhartiya D, Lenka N, eds. Pluripotent Stem Cells. Rijeka: InTech Ch. 15.
Acknowledgements
This work was supported by extramural funding from Department of Biotechnology (DBT), Government of India (Grant No.:BT/PR/14659/MED/31/108/2010) and Board of Research in Nuclear Science, Department of Atomic Energy, Government of India (Grant No.:2013/35/45/BRNS). We also acknowledge the technical support of the MSSB (Molecular Stress and Stem Cell Biology) group, School of Biotechnology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there is no conflict of interest.
Electronic supplementary material

Supplementary Figure 1
Interaction between TRF2 and CD44 proteins. A) Schematic representation of interacting amino acid residues of TRF2 and CD44 protein as analysed by Cluspro 2.0 molecular docking software. B-E) Molecular docking studies showing interaction of amino acid residues TRF2 and CD44 proteins. TRF2 amino acid residues are coded in green color, KLF4 amino acid residues are coded in pink color. F) Figure showing protein backbone of TRF2 in blue color. Direct inter residue interactions shown with solid lines and the water-mediated interactions in dash lines. Green denotes minimally frustrated regions, highly frustrated regions are in red and neutral regions are not indicated. G) Figure showing projection of local frustration distribution in the amino acid sequences of TRF2 protein. The number of contacts within 5 Å of the C-α of each residue is plotted, as classified according to their frustration index. H) Figure showing protein backbone of CD44 in blue color. Direct inter residue interactions are shown with solid lines and the water-mediated interactions in dash lines. Green denotes minimally frustrated regions, highly frustrated regions are in red and neutral regions are not shown. I) Figure showing projection of local frustration distribution in the amino acid sequences of KLF4 protein. The number of contacts within 5 Å of the C-α of each residue is plotted, as classified according to their frustration index. No interacting residues of high frustrations between TRF2 and CD44 were observed. (PNG 469 kb)
Rights and permissions
About this article
Cite this article
Saha, A., Roy, S., Kar, M. et al. Role of Telomeric TRF2 in Orosphere Formation and CSC Phenotype Maintenance Through Efficient DNA Repair Pathway and its Correlation with Recurrence in OSCC. Stem Cell Rev and Rep 14, 871–887 (2018). https://doi.org/10.1007/s12015-018-9823-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-018-9823-z