Abstract
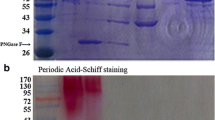
A novel GH1 β-glucosidase gene (bgla) from marine bacterium was sequenced and expressed in Escherichia coli. After purification by Ni2+ affinity chromatography, the recombinant protein was characterized. The purified recombinant enzyme showed maximum activity at 40 °C, pH 7.5 and was stable between temperatures that range from 4 to 30 °C and over the pH range of 6–10. The enzyme displayed a high tolerance to glucose and maximum stimulation at the presence of 100 mM glucose. To improve glucose tolerance of the enzyme, a site-directed mutation (f171w) was introduced into β-glucosidase. The recombinant F171W showed a higher glucose tolerance than the wild type and maintained more than 40% residual activity at the presence of 4 M glucose. Additionally, the recombinant enzymes showed notable tolerance to ethanol. These properties suggest the enzymes may have potential applications for the fermentation of lignocellulosic sugars and the production of biofuels.









Similar content being viewed by others
References
Ketudat Cairns, J. R., & Esen, A. (2010). β-Glucosidases. Cellular and Molecular Life Sciences, 67(20), 3389–3405.
Crout, D. H. G., & Vic, G. (1998). Glycosidases and glycosyl transferases in glycoside and oligosaccharide synthesis. Current Opinion in Chemical Biology, 2(1), 98–111.
Boudabbous, M., Ben, H. I., Saibi, W., Mssawra, M., Belghith, H., & Gargouri, A. (2017). Trans-glycosylation capacity of a highly glycosylated multi-specific β-glucosidase from Fusarium solani. Bioprocess and Biosystems Engineering, 40(4), 559–571.
Lundemo, P., Karlsson, E. N., & Adlercreutz, P. (2017). Eliminating hydrolytic activity without affecting the transglycosylation of a GH1 β-glucosidase. Applied Microbiology and Biotechnology, 101(3), 1121–1131.
Gilbert, H. J., Stålbrand, H., & Brumer, H. (2008). How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Current Opinion in Plant Biology, 11(3), 338–348.
Lynd, L. R., Weimer, P. J., van Zyl, W. H., & Pretorius, I. S. (2002). Microbial cellulose utilization: fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 66(3), 506–577.
Yang, F., Yang, X., Li, Z., Du, C., Wang, J., & Li, S. (2015). Overexpression and characterization of a glucose-tolerant β-glucosidase from T. aotearoense with high specific activity for cellobiose. Applied Microbiology and Biotechnology, 99(21), 8903–8915.
Dogaris, I., Mamma, D., & Kekos, D. (2013). Biotechnological production of ethanol from renewable resources by Neurospora crassa: an alternative to conventional yeast fermentations. Applied Microbiology and Biotechnology, 97(4), 1457–1473.
Wu, J., Geng, A., Xie, R., Wang, H., & Sun, J. (2018). Characterization of cold adapted and ethanol tolerant β-glucosidase from Bacillus cellulosilyticus and its application for directed hydrolysis of cellobiose to ethanol. International Journal of Biological Macromolecules, 109, 872–879.
Singhania, R. R., Patel, A. K., Sukumaran, R. K., Larroche, C., & Pandey, A. (2013). Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresource Technology, 127, 500–507.
Singh, G., Verma, A. K., & Kumar, V. (2016). Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech, 6(1), 3.
Pandey, M., & Mishra, S. (1997). Expression and characterization of Pichia etchellsii beta-glucosidase in Escherichia coli. Gene, 190(1), 45–51.
Tian, L., Liu, S., Wang, S., & Wang, L. (2016). Ligand-binding specificity and promiscuity of the main lignocellulolytic enzyme families as revealed by active-site architecture analysis. Scientific Reports, 6(1), 23605.
Tiwari P., Misra B. N., & Sangwan N. S. (2013). β-Glucosidases from the fungus Trichoderma: an efficient cellulase machinery in biotechnological applications. Biomed Research International, 2013, 203735.
Xia, W., Bai, Y., Cui, Y., Xu, X., Qian, L., Shi, P., Zhang, W., Luo, H., Zhan, X., & Yao, B. (2016). Functional diversity of family 3 β-glucosidases from thermophilic cellulolytic fungus Humicola insolens Y1. Scientific Reports, 6(1), 27062.
Rosi, I., Vinella, M., & Domizio, P. (1994). Characterization of β-glucosidase activity in yeasts of oenological origin. Journal of Applied Microbiology, 77(5), 519–527.
da Silva, R. R., da Conceição, P. J. P., de Menezes, C. L. A., de Oliveira, N. C. E., Machado, B. M., Pessoa, J. A., de Souza, G. M., da Silva, R., & Gomes, E. (2019). Biochemical characteristics and potential application of a novel ethanol and glucose-tolerant β-glucosidase secreted by Pichia guilliermondii G1.2. Journal of Biotechnology, 294, 73–80.
Sinha, S. K., & Datta, S. (2016). β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Applied Microbiology and Biotechnology, 100(19), 8399–8409.
Liew, K. J., Lim, L., Woo, H. Y., Chan, K. G., Shamsir, M. S., & Goh, K. M. (2018). Purification and characterization of a novel GH1 beta-glucosidase from Jeotgalibacillus malaysiensis. International Journal of Biological Macromolecules, 115, 1094–1102.
Fusco, F. A., Fiorentino, G., Pedone, E., Contursi, P., Bartolucci, S., & Limauro, D. (2018). Biochemical characterization of a novel thermostable β-glucosidase from Dictyoglomus turgidum. International Journal of Biological Macromolecules, 113, 783–791.
Zhong, F. L., Ma, R., Jiang, M., Dong, W. W., Jiang, J., Wu, S., Li, D., & Quan, L. H. (2016). Cloning and characterization of ginsenoside-hydrolyzing β-glucosidase from Lactobacillus brevis that transforms ginsenosides Rb1 and F2 into ginsenoside Rd and compound K. Journal of Microbiology and Biotechnology, 26(10), 1661–1667.
Shipkowski, S., & Brenchley, J. E. (2005). Characterization of an unusual cold-active beta-glucosidase belonging to family 3 of the glycoside hydrolases from the psychrophilic isolate Paenibacillus sp. strain C7. Applied and Environmental Microbiology, 71(8), 4225–4232.
Cao, L. C., Wang, Z. J., Ren, G. H., Kong, W., Li, L., Xie, W., & Liu, Y. H. (2015). Engineering a novel glucose-tolerant β-glucosidase as supplementation to enhance the hydrolysis of sugarcane bagasse at high glucose concentration. Biotechnology for Biofuels, 8(1), 202.
Pang, P., Cao, L. C., Liu, Y. H., Xie, W., & Wang, Z. (2017). Structures of a glucose-tolerant β-glucosidase provide insights into its mechanism. Journal of Structural Biology, 198(3), 154–162.
Matsuzawa, T., Jo, T., Uchiyama, T., Manninen, J. A., Arakawa, T., Miyazaki, K., Fushinobu, S., & Yaoi, K. (2016). Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic β-glucosidase Td2F2. FEBS Journal, 283(12), 2340–2353.
Salgado, J. C. S., Meleiro, L. P., Carli, S., & Ward, R. J. (2018). Glucose tolerant and glucose stimulated β-glucosidases—a review. Bioresource Technology, 267, 704–713.
de Giuseppe P. O., Souza T. A. C. B., Souza F. H. M., Zanphorlin L. M., Machado C. B., Ward R. J., Jorge J. A., Furriel R. P. M., & Murakami M. T. (2014). Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Crystallographica. Section D: Biological Crystallography, 70(Pt 6), 1631–1639.
Guo, B., Amano, Y., & Nozaki, K. (2016). Improvements in glucose sensitivity and stability of Trichoderma reesei β-glucosidase using site-directed mutagenesis. PLoS One, 11(1), e0147301.
Yang, Y., Zhang, X., Yin, Q., Fang, W., Fang, Z., Wang, X., Zhang, X., & Xiao, Y. (2015). A mechanism of glucose tolerance and stimulation of GH1 β-glucosidases. Scientific Reports, 5(1), 17296.
Sun, J., Wang, W., Yao, C., Dai, F., Zhu, X., Liu, J., & Hao, J. (2018). Overexpression and characterization of a novel cold-adapted and salt-tolerant GH1 β-glucosidase from the marine bacterium Alteromonas sp. L82. Journal of Microbiology, 56(9), 656–664.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). Swiss-model: an automated protein homology-modeling server. Nucleic Acids Research, 31(1), 3381–3385.
Morris, G. M., Goodsell, D., Halliday, R., Huey, R., Hart, W., Belew, R. K., & Olson, A. J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry, 19(14), 1639–1662.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). Autodock4 and autodocktools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791.
Fang, Z., Fang, W., Liu, J., Hong, Y., Peng, H., Zhang, X., Sun, B., & Xiao, Y. (2010). Cloning and characterization of a β-glucosidase from marine microbial metagenome with excellent glucose tolerance. Journal of Microbiology and Biotechnology, 20(9), 1351–1358.
Souza, F. H. M., Nascimento, C. V., Rosa, J. C., Masui, D. C., Leone, F. A., Jorge, J. A., & Furriel, R. P. M. (2010). Purification and biochemical characterization of a mycelial glucose- and xylose-stimulated β-glucosidase from the thermophilic fungus Humicola insolens. Process Biochemistry, 45(2), 272–278.
Lee, H. L., Chang, C. K., Jeng, W. Y., Wang, A. H., & Liang, P. H. (2012). Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Engineering, Design and Selection, 25(11), 733–740.
Crespim, E., Zanphorlin, L. M., de Souza, F. H. M., Diogo, J. A., Gazolla, A. C., Machado, C. B., Figueiredo, F., Sousa, A. S., Nóbrega, F., Pellizari, V. H., Murakami, M. T., & Ruller, R. (2016). A novel cold-adapted and glucose-tolerant GH1 β-glucosidase from Exiguobacterium antarcticum B7. International Journal of Biological Macromolecules, 82, 375–380.
Geiger, G., Furrer, G., Funk, F., Brandl, H., & Schulin, R. (1999). Heavy metal effects on beta-glucosidase activity influenced by pH and buffer systems. Journal of Enzyme Inhibition and Medicinal Chemistry, 14(5), 365–379.
Mai, Z., Yang, J., Tian, X., Li, J., & Zhang, S. (2013). Gene cloning and characterization of a novel salt-tolerant and glucose-enhanced β-glucosidase from a marine streptomycete. Applied Biochemistry and Biotechnology, 169(5), 1512–1522.
Fang, W., Yang, Y., Zhang, X., Yin, Q., Zhang, X., Wang, X., Fang, Z., & Xiao, Y. (2016). Improve ethanol tolerance of β-glucosidase Bgl1A by semi-rational engineering for the hydrolysis of soybean isoflavone glycosides. Journal of Biotechnology, 227, 64–71.
Lu, J., Du, L., Wei, Y., Hu, Y., & Huang, R. (2013). Expression and characterization of a novel highly glucose-tolerant β-glucosidase from a soil metagenome. Acta Biochimica et Biophysica Sinica, 45(8), 664–673.
Wierzbicka-Woś, A., Bartasun, P., Cieśliński, H., & Kur, J. (2013). Cloning and characterization of a novel cold-active glycoside hydrolase family 1 enzyme with β-glucosidase, β-fucosidase and β-galactosidase activities. BMC Biotechnology, 13(1), 22.
Meleiro, L. P., Salgado, J. C. S., Maldonado, R. F., Alponti, J. S., Zimbardi, A. L. R. L., Jorge, J. A., Ward, R. J., & Furriel, R. P. M. (2015). A Neurospora crassa β-glucosidase with potential for lignocellulose hydrolysis shows strong glucose tolerance and stimulation by glucose and xylose. Journal of Molecular Catalysis B: Enzymatic, 122, 131–140.
Chamoli, S., Kumar, P., Navani, N. K., & Verma, A. K. (2016). Secretory expression, characterization and docking study of glucose-tolerant β-glucosidase from B. subtilis. International Journal of Biological Macromolecules, 85, 425–433.
Funding
This study was funded by the National Natural Science Foundation of China (31900035); the Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022018006); the Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO.2020TD67); the Financial Fund of the Ministry of Agriculture, P. R. China (No.NFZX2013); and the Scientific and the Technological Innovation Project Financially Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2016ASKJ14).
Author information
Authors and Affiliations
Contributions
J. Sun and J. Hao contributed to the conception and design of the study; J. Sun contributed to the Data curation; J. Sun, J. Hao, and W. Wang contributed significantly to analysis and manuscript preparation; J. Sun and Y. Ying performed writing-review and editing.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Statement
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 417 kb)
Rights and permissions
About this article
Cite this article
Sun, J., Wang, W., Ying, Y. et al. A Novel Glucose-Tolerant GH1 β-Glucosidase and Improvement of Its Glucose Tolerance Using Site-Directed Mutation. Appl Biochem Biotechnol 192, 999–1015 (2020). https://doi.org/10.1007/s12010-020-03373-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03373-z