Abstract
Purpose of Review
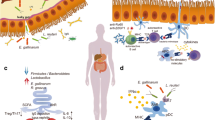
The resident gut microbiota serves as a double-edged sword that aids the host in multiple ways to preserve a healthy equilibrium and serve as early companions and boosters for the gradual evolution of our immune defensive layers; nevertheless, the perturbation of the symbiotic resident intestinal communities has a profound impact on autoimmunity induction, particularly in systemic lupus erythematosus (SLE). Herein, we seek to critically evaluate the microbiome research in SLE with a focus on intestinal dysbiosis.
Recent Findings
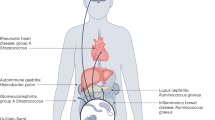
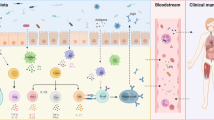
SLE is a complex and heterogeneous disorder with self-attack due to loss of tolerance, and there is aberrant excessive immune system activation. There is mounting evidence suggesting that intestinal flora disturbances may accelerate the formation and progression of SLE, presumably through a variety of mechanisms, including intestinal barrier dysfunction and leaky gut, molecular mimicry, bystander activation, epitope spreading, gender bias, and biofilms.
Summary
Gut microbiome plays a critical role in SLE pathogenesis, and additional studies are warranted to properly define the impact of gut microbiome in SLE, which can eventually lead to new and potentially safer management approaches for this debilitating disease.


Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med. 2020;172(11):ITC81–96. https://doi.org/10.7326/aitc202006020.
Durcan L, O’Dwyer T, Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393(10188):2332–43. https://doi.org/10.1016/s0140-6736(19)30237-5.
Agmon-Levin N, Mosca M, Petri M, Shoenfeld Y. Systemic lupus erythematosus one disease or many? Autoimmun Rev. 2012;11(8):593–5. https://doi.org/10.1016/j.autrev.2011.10.020.
Kaul A, Gordon C, Crow MK, Touma Z, Urowitz MB, van Vollenhoven R, et al. Systemic lupus erythematosus. Nature Reviews Disease Primers. 2016;2. https://doi.org/10.1038/nrdp.2016.39.
Bengtsson AA, Rönnblom L. Role of interferons in SLE. Best Pract Res Clin Rheumatol. 2017;31(3):415–28. https://doi.org/10.1016/j.berh.2017.10.003.
Silverman GJ. The microbiome in SLE pathogenesis. Nat Rev Rheumatol. 2019;15(2):72–4. https://doi.org/10.1038/s41584-018-0152-z.
Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol. 2020;21(6):605–14. https://doi.org/10.1038/s41590-020-0677-6.
Tsokos GC, Lo MS, Reis PC, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016;12(12):716–30. https://doi.org/10.1038/nrrheum.2016.186.
Zhang LS, Qing PY, Yang H, Wu YK, Liu Y, Luo YB. Gut microbiome and metabolites in systemic lupus erythematosus: link, mechanisms and intervention. Frontiers in Immunology. 2021;12. https://doi.org/10.3389/fimmu.2021.686501.
Borchers AT, Keen CL, Shoenfeld Y, Gershwin ME. Surviving the butterfly and the wolf: mortality trends in systemic lupus erythematosus. Autoimmun Rev. 2004;3(6):423–53. https://doi.org/10.1016/j.autrev.2004.04.002.
Bernatsky S, Boivin JF, Joseph L, Manzi S, Ginzler E, Gladman DD, et al. Mortality in systemic lupus erythematosus. Arthritis Rheum. 2006;54(8):2550–7. https://doi.org/10.1002/art.21955.
Jorge AM, Lu N, Zhang YQ, Rai SK, Choi HK. Unchanging premature mortality trends in systemic lupus erythematosus: a general population-based study (1999–2014). Rheumatology. 2018;57(2):337–44. https://doi.org/10.1093/rheumatology/kex412.
Xiang ST, Qu YQ, Qian SH, Wang RY, Wang Y, Jin YB, et al. Association between systemic lupus erythematosus and disruption of gut microbiota: a meta-analysis. Lupus Sci Med. 2022;9(1):e000599. https://doi.org/10.1136/lupus-2021-000599.
De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2019;195(1):74–85. https://doi.org/10.1111/cei.13158.
Saadat YR, Hejazian M, Bastami M, Khatibi SMH, Ardalan M, Vahed SZ. The role of microbiota in the pathogenesis of lupus: dose it impact lupus nephritis? Pharmacol Res. 2019;139:191–8. https://doi.org/10.1016/j.phrs.2018.11.023.
Yaigoub H, Fath N, Tirichen H, Wu CX, Li RS, Li YF. Bidirectional crosstalk between dysbiotic gut microbiota and systemic lupus erythematosus: what is new in therapeutic approaches? Clinical Immunol. 2022;244:109109. https://doi.org/10.1016/j.clim.2022.109109.
Zhang HS, Liao XF, Sparks JB. Luo XM Dynamics of gut microbiota in autoimmune lupus applied and environmental microbiology. Appl Environ Microbiol. 2014;80(24):7551–60. https://doi.org/10.1128/aem.02676-14. The intestinal dysbiosis in lupus was reported for the first time in a murine model.
Kim JW, Kwok SK, Choe JY, Park SH. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int J Mol Sci. 2019;20(19):4871. https://doi.org/10.3390/ijms20194871.
Silverman GJ, Azzouz DF, Alekseyenko AV. Systemic lupus erythematosus and dysbiosis in the microbiome: cause or effect or both? Curr Opin Immunol. 2019;61:80–5. https://doi.org/10.1016/j.coi.2019.08.007.
Neuman H, Koren O. The gut microbiota: a possible factor influencing systemic lupus erythematosus. Curr Opin Rheumatol. 2017;29(4):374–7. https://doi.org/10.1097/bor.0000000000000395.
Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. Mbio. 2014;5(5). https://doi.org/10.1128/mBio.01548-14. For the first time, next-generation sequencing approaches are used to characterize intestinal dysbiosis linked to SLE.
Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–9. https://doi.org/10.1126/science.1124234.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. https://doi.org/10.1038/nri2515.
Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59-U70. https://doi.org/10.1038/nature08821.
Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76(3):473–93. https://doi.org/10.1007/s00018-018-2943-4.
Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489(7415):250–6. https://doi.org/10.1038/nature11553.
Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68(4):669. https://doi.org/10.1128/mmbr.68.4.669-685.2004.
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. https://doi.org/10.1038/nature11550.
Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31(1):107–33. https://doi.org/10.1146/annurev.mi.31.100177.000543.
Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4(6):478–85. https://doi.org/10.1038/nri1373.
Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–40. https://doi.org/10.1016/j.cell.2016.01.013.
Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–35. https://doi.org/10.1038/nri3430.
Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–20. https://doi.org/10.1126/science.1104816.
Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9(10):577–89. https://doi.org/10.1038/nrgastro.2012.156.
Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10(3):159–69. https://doi.org/10.1038/nri2710.
Rosser EC, Mauri C. A clinical update on the significance of the gut microbiota in systemic autoimmunity. J Autoimmun. 2016;74:85–93. https://doi.org/10.1016/j.jaut.2016.06.009.
Biedermann L, Rogler G. The intestinal microbiota: its role in health and disease. Eur J Pediatr. 2015;174(2):151–67. https://doi.org/10.1007/s00431-014-2476-2.
Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biologics-Targets & Therapy. 2011;5:71–86. https://doi.org/10.2147/btt.S19099.
Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22(10):1079–89. https://doi.org/10.1038/nm.4185.
Lange K, Buerger M, Stallmach A, Bruns T. Effects of antibiotics on gut microbiota. Dig Dis. 2016;34(3):260–8. https://doi.org/10.1159/000443360.
Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int J Mol Sci. 2020;21(1):107. https://doi.org/10.3390/ijms21010107.
Rosenbaum JT, Silverman GJ. The microbiome and systemic lupus erythematosus. N Engl J Med. 2018;378(23):2236–7. https://doi.org/10.1056/NEJMcibr1804368.
Shamriz O, Mizrahi H, Werbner M, Shoenfeld Y, Avni O, Koren O. Microbiota at the crossroads of autoimmunity. Autoimmun Rev. 2016;15(9):859–69. https://doi.org/10.1016/j.autrev.2016.07.012.
Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787–803. https://doi.org/10.3748/wjg.v21.i29.8787.
Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. https://doi.org/10.1016/j.cell.2014.03.011.
Chung HC, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–93. https://doi.org/10.1016/j.cell.2012.04.037.
Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–99. https://doi.org/10.1016/j.cell.2014.09.053.
Tezuka H, Abe Y, Asano J, Sato T, Liu JJ, Iwata M, et al. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity. 2011;34(2):247–57. https://doi.org/10.1016/j.immuni.2011.02.002.
Sterlin D, Fadlallah J, Slack E, Gorochov G. The antibody/microbiota interface in health and disease. Mucosal Immunol. 2020;13(1):3–11. https://doi.org/10.1038/s41385-019-0192-y.
Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–32. https://doi.org/10.1038/nri.2017.7.
Olesen SW, Alm EJ. Dysbiosis is not an answer. Nat Microbiol. 2016;1(12):16228. https://doi.org/10.1038/nmicrobiol.2016.228.
Christovich A, Luo XM. Gut microbiota, leaky gut, and autoimmune diseases. Frontiers in Immunology. 2022;13. https://doi.org/10.3389/fimmu.2022.946248.
Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–70. https://doi.org/10.1126/science.1240527.
Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592. https://doi.org/10.1126/science.aah3648.
Tang WHW, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120(7):1183–96. https://doi.org/10.1161/circresaha.117.309715.
Wang ZN, Zhao YZ. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. 2018;9(5):416–31. https://doi.org/10.1007/s13238-018-0549-0.
Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Medicine. 2016;8. https://doi.org/10.1186/s13073-016-0303-2.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480-U7. https://doi.org/10.1038/nature07540.
Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30(6):787–97. https://doi.org/10.1177/0884533615609896.
Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17(5):592–602. https://doi.org/10.1016/j.chom.2015.04.007.
Fujimura KE, Sitarik AR, Haystad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187–91. https://doi.org/10.1038/nm.4176.
Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic hiv infection. J Infect Dis. 2015;211(1):19–27. https://doi.org/10.1093/infdis/jiu409.
Zhuang ZQ, Shen LL, Li WW, Fu X, Zeng F, Gui L, et al. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis. 2018;63(4):1337–46. https://doi.org/10.3233/jad-180176.
Petra AI, Panagiotidou S, Hatziagelaki E, Stewart JM, Conti P, Theoharides TC. Gut-microbiota-brain axis and its effect on neuropsychiatric disorders with suspected immune dysregulation. Clin Ther. 2015;37(5):984–95. https://doi.org/10.1016/j.clinthera.2015.04.002.
Ozen S, Lutz HL, Rivera VM, Reiff A, Batu ED, Anderson E, et al. Microbiome is not linked to clinical disease severity of familial Mediterranean fever in an international cohort of children. Clin Exp Rheumatol. 2021;39(5):102–8. https://doi.org/10.55563/clinexprheumatol/olvbyd.
Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA. 2017;114(40):10713–8. https://doi.org/10.1073/pnas.1711235114.
Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Scientific Reports. 2016;6. https://doi.org/10.1038/srep28484.
Cao Y, Oh J, Xue M, Huh WJ, Wang J, Gonzalez-Hernandez JA, et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science. 2022;378(6618):eabm3233. https://doi.org/10.1126/science.abm3233.
Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser H, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. https://doi.org/10.1038/s41564-018-0306-4.
Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. Bmc Medicine. 2013;11. https://doi.org/10.1186/1741-7015-11-46.
Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583. https://doi.org/10.1038/s41586-018-0617-x.
Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2. https://doi.org/10.7554/eLife.01202.
Zhang X, Zhang DY, Jia HJ, Feng Q, Wang DH, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. https://doi.org/10.1038/nm.3914.
Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. Pathogenic autoreactive T and B cells cross-react with mimotopes expressed by a common human gut commensal to trigger autoimmunity. Cell Host Microbe. 2019;26(1):100-13 e8. https://doi.org/10.1016/j.chom.2019.05.003.
Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. 2019;78(7):947–56. https://doi.org/10.1136/annrheumdis-2018-214856.
Lin L, Zhang JQ. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. Bmc Immunology. 2017;18. https://doi.org/10.1186/s12865-016-0187-3.
Posadas-Romero C, Torres-Tamayo M, Zamora-Gonzalez J, Aguilar-Herrera BE, Posadas-Sanchez R, Cardoso-Saldana G, et al. High insulin levels and increased low-density lipoprotein oxidizability in pediatric patients with systemic lupus erythematosus. Arthritis Rheum. 2004;50(1):160–5. https://doi.org/10.1002/art.11472.
Yan B, Huang J, Zhang C, Hu X, Gao M, Shi A, et al. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod Rheumatol. 2016;26(6):914–22. https://doi.org/10.3109/14397595.2016.1158895.
Wu TF, Xie C, Han J, Ye YJ, Weiel J, Li Q, et al. Metabolic disturbances associated with systemic lupus erythematosus. Plos One. 2012;7(6). https://doi.org/10.1371/journal.pone.0037210.
Borba EF, Bonfa E. Dyslipoproteinemias in systemic lupus erythematosus: influence of disease, activity, and anticardiolipin antibodies. Lupus. 1997;6(6):533–9. https://doi.org/10.1177/096120339700600610.
He JQ, Chan TL, Hong XP, Zheng FP, Zhu CX, Yin LH, et al. Microbiome and metabolome analyses reveal the disruption of lipid metabolism in systemic lupus erythematosus. Frontiers in Immunology. 2020;11. https://doi.org/10.3389/fimmu.2020.01703.
Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5. https://doi.org/10.1038/nature12726.
Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic T<sub>reg</sub> cell homeostasiS. Science. 2013;341(6145):569–73. https://doi.org/10.1126/science.1241165.
Rodriguez-Carrio J, Lopez P, Sanchez B, Gonzalez S, Gueimonde M, Margolles A, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Frontiers in Immunology. 2017;8. https://doi.org/10.3389/fimmu.2017.00023.
Choi S-C, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Trans Med. 2020;12(551):eaax2220. https://doi.org/10.1126/scitranslmed.aax2220.
He Z, Shao T, Li H, Xie Z, Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016;8:64. https://doi.org/10.1186/s13099-016-0146-9.
Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med. 2018;10(434). https://doi.org/10.1126/scitranslmed.aan2306. This study suggested that Ro60 ortholog-producing commensal species may initiate and also maintain autoimmunity in lupus in genetically predisposed individuals via a molecular mimicry mechanism.
Li Y, Wang HF, Li X, Li HX, Zhang Q, Zhou HW, et al. Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin Sci. 2019;133(7):821–38. https://doi.org/10.1042/cs20180841.
Zhang SX, Wang J, Chen JW, Zhang MX, Zhang YF, Hu FY, et al. The level of peripheral regulatory T cells is linked to changes in gut commensal microflora in patients with systemic lupus erythematosus. Ann Rheum Dis. 2021;80(11):e177. https://doi.org/10.1136/annrheumdis-2019-216504.
van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary Sjögren’s syndrome and systemic lupus erythematosus. J Autoimmun. 2019;97:77–87. https://doi.org/10.1016/j.jaut.2018.10.009.
Guo M, Wang H, Xu S, Zhuang Y, An J, Su C, et al. Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes. 2020;11(6):1758–73. https://doi.org/10.1080/19490976.2020.1768644.
López P, de Paz B, Rodríguez-Carrio J, Hevia A, Sánchez B, Margolles A, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep. 2016;6:24072. https://doi.org/10.1038/srep24072.
Gerges MA, Esmaeel NE, Makram WK, Sharaf DM, Gebriel MG. Altered profile of fecal microbiota in newly diagnosed systemic lupus erythematosus Egyptian Patients. Int J Microbiol. 2021;2021:9934533. https://doi.org/10.1155/2021/9934533.
Toumi E, Goutorbe B, Plauzolles A, Bonnet M, Mezouar S, Militello M, et al. Gut microbiota in systemic lupus erythematosus patients and lupus mouse model: a cross species comparative analysis for biomarker discovery. Front Immunol. 2022;13:943241. https://doi.org/10.3389/fimmu.2022.943241.
Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol. 2018;84(4). https://doi.org/10.1128/aem.02288-17.
Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, et al. The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12(5). https://doi.org/10.3390/nu12051474.
Man SM, Kaakoush NO, Mitchell HM. The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat Rev Gastroenterol Hepatol. 2011;8(3):152–68. https://doi.org/10.1038/nrgastro.2011.3.
Wei F, Xu HF, Yan CX, Rong CL, Liu BY, Zhou HZ. Changes of intestinal flora in patients with systemic lupus erythematosus in northeast China. Plos One. 2019;14(3). https://doi.org/10.1371/journal.pone.0213063.
Bellocchi C, Fernández-Ochoa Á, Montanelli G, Vigone B, Santaniello A, Quirantes-Piné R, et al. Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J Clin Med. 2019;8(9). https://doi.org/10.3390/jcm8091291.
Wen M, Liu T, Zhao M, Dang X, Feng S, Ding X, et al. Correlation analysis between gut microbiota and metabolites in children with systemic lupus erythematosus. J Immunol Res. 2021;2021:5579608. https://doi.org/10.1155/2021/5579608.
Zegarra-Ruiz DF, El Beidaq A, Iñiguez AJ, Di Ricco ML, Vieira SM, Ruff WE, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25(1):113–276. https://doi.org/10.1016/j.chom.2018.11.009.
Bankole A, Luo X, Husen Z. A comparative analysis of gut microbiota between systemic lupus erythematosus patients and non-autoimmune controls a single centrecenter cohort experience. Lupus Sci Med. 2017;4(Suppl 1):A155–6. https://doi.org/10.1136/lupus-2017-000215.354.
Chen BD, Jia XM, Xu JY, Zhao LD, Ji JY, Wu BX, et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol. 2021;73(2):232–43. https://doi.org/10.1002/art.41511. This research study offers comprehensive insights into the microbiota composition of individuals with untreated SLE by examining the functional characteristics of the microbiota and the identification of peptides that mimic autoantigens.
Bunker JJ, Drees C, Watson AR, Plunkett CH, Nagler CR, Schneewind O, et al. B cell superantigens in the human intestinal microbiota. Sci Transl Med. 2019;11(507). https://doi.org/10.1126/scitranslmed.aau9356.
Pan Q, Guo F, Huang Y, Li A, Chen S, Chen J, et al. Gut microbiota dysbiosis in systemic lupus erythematosus: novel insights into mechanisms and promising therapeutic strategies. Front Immunol. 2021;12:799788. https://doi.org/10.3389/fimmu.2021.799788.
Rojo D, Hevia A, Bargiela R, López P, Cuervo A, González S, et al. Ranking the impact of human health disorders on gut metabolism: systemic lupus erythematosus and obesity as study cases. Sci Rep. 2015;5:8310. https://doi.org/10.1038/srep08310.
Ma YYZ, Guo RR, Sun YD, Li X, He L, Li Z, et al. Lupus gut microbiota transplants cause autoimmunity and inflammation. Clin Immunol. 2021;233:108892. https://doi.org/10.1016/j.clim.2021.108892.
Tomofuji Y, Maeda Y, Oguro-Igashira E, Kishikawa T, Yamamoto K, Sonehara K, et al. Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese. Ann Rheum Dis. 2021;80(12):1575–83. https://doi.org/10.1136/annrheumdis-2021-220687.
Tomofuji Y, Kishikawa T, Maeda Y, Ogawa K, Nii T, Okuno T, et al. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Ann Rheum Dis. 2022;81(2):278–88. https://doi.org/10.1136/annrheumdis-2021-221267.
Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5(1):73. https://doi.org/10.1186/s40168-017-0300-8.
Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med. 2020;12(551). https://doi.org/10.1126/scitranslmed.aax2220.
Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol Med. 2019;25(1):35. https://doi.org/10.1186/s10020-019-0102-5.
Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359(6380):1156–61. https://doi.org/10.1126/science.aar7201.
López P, Sánchez B, Margolles A, Suárez A. Intestinal dysbiosis in systemic lupus erythematosus: cause or consequence? Curr Opin Rheumatol. 2016;28(5):515–22. https://doi.org/10.1097/bor.0000000000000309.
Chen YF, Lin J, Xiao LL, Zhang X, Zhao LD, Wang M, et al. Gut microbiota in systemic lupus erythematosus: a fuse and a solution. J Autoimmun. 2022;132:102867. https://doi.org/10.1016/j.jaut.2022.102867.
Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50(8):1–9. https://doi.org/10.1038/s12276-018-0126-x.
Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68(8):1516–26. https://doi.org/10.1136/gutjnl-2019-318427.
Charoensappakit A, Sae-khow K, Leelahavanichkul A. Gut barrier damage and gut translocation of pathogen molecules in lupus, an impact of innate immunity (macrophages and neutrophils) in autoimmune disease. Int J Mol Sci. 2022;23(15):8223. https://doi.org/10.3390/ijms23158223.
Toral M, Robles-Vera I, Romero M, de la Visitación N, Sánchez M, O’Valle F, et al. Lactobacillus fermentum CECT5716: a novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. FASEB J. 2019;33(9):10005–18. https://doi.org/10.1096/fj.201900545RR.
Silverman GJ, Deng J, Azzouz DF. Sex-dependent Lupus Blautia (Ruminococcus) gnavus strain induction of zonulin-mediated intestinal permeability and autoimmunity. Frontiers in Immunology. 2022;13. https://doi.org/10.3389/fimmu.2022.897971.
Mu QH, Kirby J, Reilly CM, Luo XM. Leaky gut as a danger signal for autoimmune diseases. frontiers in immunology. 2017;8. https://doi.org/10.3389/fimmu.2017.00598.
Haga HJ, Brun JG, Berntzen HB, Cervera R, Khamashta M, Hughes GRV. Calprotectin in patients with systemic lupus-erythematosus - relation to clinical and laboratory parameters of disease-activity. Lupus. 1993;2(1):47–50. https://doi.org/10.1177/096120339300200108.
Gisbert JP, Bermejo F, Perez-Calle JL, Taxonera C, Vera I, McNicholl AG, et al. Fecal Calprotectin and lactoferrin for the prediction of inflammatory bowel disease relapse. Inflamm Bowel Dis. 2009;15(8):1190–8. https://doi.org/10.1002/ibd.20933.
Liu B, Yang Y, Dai J, Medzhitov R, Freudenberg MA, Zhang PL, et al. TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J Immunol. 2006;177(10):6880–8. https://doi.org/10.4049/jimmunol.177.10.6880.
Lee TP, Tang SJ, Wu MF, Song YC, Yu CL, Sun KH. Transgenic overexpression of anti-double-stranded DNA autoantibody and activation of Toll-like receptor 4 in mice induce severe systemic lupus erythematosus syndromes. J Autoimmun. 2010;35(4):358–67. https://doi.org/10.1016/j.jaut.2010.07.007.
Nockher WA, Wigand R, Schoeppe W, Scherberich JE. Elevated levels of soluble cd14 in serum of patients with systemic lupus-erythematosus. Clin Exp Immunol. 1994;96(1):15–9.
Mu QH, Zhang HS, Luo XM. SLE: Another autoimmune disorder influenced by microbes and diet? Frontiers in Immunology. 2015;6. https://doi.org/10.3389/fimmu.2015.00608.
Shi L, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One. 2014;9(5):e93846. https://doi.org/10.1371/journal.pone.0093846.
Thim-uam A, Surawut S, Issara-Amphorn J, Jaroonwitchawan T, Hiengrach P, Chatthanathon P, et al. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Scientific Reports. 2020;10(1). https://doi.org/10.1038/s41598-019-57275-0.
Sturgeon C, Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers. 2016;4(4):e1251384. https://doi.org/10.1080/21688370.2016.1251384.
Blank M, Barzilai O, Shoenfeld Y. Molecular mimicry and auto-immunity. Clin Rev Allergy Immunol. 2007;32(1):111–8. https://doi.org/10.1007/BF02686087.
Shoenfeld Y, Vilner Y, Coates AR, Rauch J, Lavie G, Shaul D, et al. Monoclonal anti-tuberculosis antibodies react with DNA, and monoclonal anti-DNA autoantibodies react with Mycobacterium tuberculosis. Clin Exp Immunol. 1986;66(2):255–61.
Cervera R, Piette JC, Font J, Khamashta MA, Cervera R, Piette JC, et al. Antiphospholipid syndrome - Clinical and immunologic manifestations and patterns of disease expression in a cohort of 1,000 patients. Arthritis Rheum. 2002;46(4):1019–27. https://doi.org/10.1002/art.10187.
James JA, Robertson JM. Lupus and Epstein-Barr. Curr Opin Rheumatol. 2012;24(4):383–8. https://doi.org/10.1097/BOR.0b013e3283535801.
Vanderlugt CJ, Miller SD. Epitope spreading. Curr Opin Immunol. 1996;8(6):831–6. https://doi.org/10.1016/s0952-7915(96)80012-4.
Vanderlugt CL, Miller SD. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat Rev Immunol. 2002;2(2):85–95. https://doi.org/10.1038/nri724.
Deshmukh US, Lewis JE, Gaskin F, Kannapell CC, Waters ST, Lou YH, et al. Immune responses to Ro60 and its peptides in mice. I. The nature of the immunogen and endogenous autoantigen determine the specificities of the induced autoantibodies. J Exp Med. 1999;189(3):531–40. https://doi.org/10.1084/jem.189.3.531.
Poole BD, Scofield RH, Harley JB, James JA. Epstein-Barr virus and molecular mimicry in systemic lupus erythematosus. Autoimmunity. 2006;39(1):63–70. https://doi.org/10.1080/08916930500484849.
Pacheco Y, Acosta-Ampudia Y, Monsalve DM, Chang C, Gershwin ME, Anaya JM. Bystander activation and autoimmunity. J Autoimmun. 2019;103:102301. https://doi.org/10.1016/j.jaut.2019.06.012.
Lee H-G, Cho M-Z, Choi J-M. Bystander CD4+ T cells: crossroads between innate and adaptive immunity. Exp Mol Med. 2020;52(8):1255–63. https://doi.org/10.1038/s12276-020-00486-7.
Tough DF, Sun SQ, Sprent J. T cell stimulation in vivo by lipopolysaccharide (LPS). J Exp Med. 1997;185(12):2089–94. https://doi.org/10.1084/jem.185.12.2089.
Zandman-Goddard G, Peeva E, Shoenfeld Y. Gender and autoimmunity. Autoimmun Rev. 2007;6(6):366–72. https://doi.org/10.1016/j.autrev.2006.10.001.
Peeva E, Zouali M. Spotlight on the role of hormonal factors in the emergence of autoreactive B-lymphocytes. Immunol Lett. 2005;101(2):123–43. https://doi.org/10.1016/j.imlet.2005.05.014.
Grimaldi CM, Hill L, Xu X, Peeva E, Diamond B. Hormonal modulation of B cell development and repertoire selection. Mol Immunol. 2005;42(7):811–20. https://doi.org/10.1016/j.molimm.2004.05.014.
Hughes GC, Choubey D. Modulation of autoimmune rheumatic diseases by oestrogen and progesterone. Nat Rev Rheumatol. 2014;10(12):740–51. https://doi.org/10.1038/nrrheum.2014.144.
Johnson BM, Gaudreau MC, Gudi R, Brown R, Gilkeson G, Vasu C. Gut microbiota differently contributes to intestinal immune phenotype and systemic autoimmune progression in female and male lupus-prone mice. J Autoimmun. 2020;108:102420. https://doi.org/10.1016/j.jaut.2020.102420.
Mu Q, Cabana-Puig X, Mao J, Swartwout B, Abdelhamid L, Cecere TE, et al. Pregnancy and lactation interfere with the response of autoimmunity to modulation of gut microbiota. Microbiome. 2019;7(1):105. https://doi.org/10.1186/s40168-019-0720-8.
Edwards MR, Dai R, Heid B, Cecere TE, Khan D, Mu Q, et al. Commercial rodent diets differentially regulate autoimmune glomerulonephritis, epigenetics and microbiota in MRL/lpr mice. Int Immunol. 2017;29(6):263–76. https://doi.org/10.1093/intimm/dxx033.
Costerton JW. Introduction to biofilm. Int J Antimicrob Agents. 1999;11(3):217–21. https://doi.org/10.1016/S0924-8579(99)00018-7.
Gallo PM, Rapsinski GJ, Wilson RP, Oppong GO, Sriram U, Goulian M, et al. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity. 2015;42(6):1171–84. https://doi.org/10.1016/j.immuni.2015.06.002.
Acknowledgements
We appreciate Dr. Hande Canpinar’s kind feedback on this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Competing Interests
S.O. has received consultancy fees form Novartis and SOBI, both unrelated to the presented work. F.N.C.K. declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kalayci, F.N.C., Ozen, S. Possible Role of Dysbiosis of the Gut Microbiome in SLE. Curr Rheumatol Rep 25, 247–258 (2023). https://doi.org/10.1007/s11926-023-01115-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11926-023-01115-8