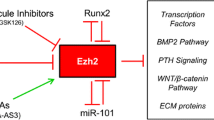
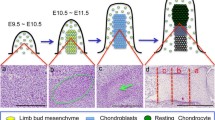
Abstract
Purpose of Review
Epigenetic regulation is a distinct mechanism of gene regulation that functions by modulating chromatin structure and accessibility. Polycomb Repressive Complex 2 (PRC2) is a conserved chromatin regulator that is required in the developing embryo to control the expression of key developmental genes. An emerging feature of PRC2 is that it not only allows for binary ON/OFF states of gene expression but can also modulate gene expression in feed-forward loops to change the outcome of gene regulatory networks. This striking feature of epigenetic modulation has improved our understanding of musculoskeletal development.
Recent Findings
Recent advances in mouse embryos unravel a range of phenotypes that demonstrate the tissue-specific, temporal, and spatial role of PRC2 during organogenesis and cell fate decisions in vivo. Here, we take a detailed view of how PRC2 functions during the development of calvarial bone and skin.
Summary
Based on the emerging evidence, we propose that PRC2 serves as a “dimmer switch” to modulate gene expression of target genes by altering the expression of activators and inhibitors. This review highlights the findings from contemporary research that allow us to investigate the unique developmental potential of intramembranous calvarial bones.




Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905.
Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell. 2010;19:651–61.
Margueron REL, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9.
Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208.
Su I-H, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–31.
Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–64.
Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–71.
Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–68.
Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, et al. Mutations in EZH2 cause weaver syndrome. Am J Hum Genet. 2012;90:110–8.
Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte SDV, Ramsay E, et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–33.
Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–46.
•• Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, et al. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. 2017;355:eaal2913.
Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017;171:34–57.
•• Bracken AP, Brien GL, Verrijzer CP. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev. 2019;33:936–59.
Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–51.
Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–9.
Yu J-R, Lee C-H, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33:903–35.
Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci. 2010;107:20980–5.
Zee BM, Britton L-MP, Wolle D, Haberman DM, Garcia BA. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol. 2012;32:2503–14.
Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev Cold Spring Harbor Lab. 2010;24:368–80.
Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature Nature Publishing Group. 2010;464:306–10.
Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–31.
Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–50.
Perino M, van Mierlo G, Karemaker ID, van Genesen S, Vermeulen M, Marks H, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet Nature Publishing Group. 2018;50:1002–10.
Youmans DT, Schmidt JC, Cech TR. Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 2018;32:794–805.
Kasinath V, Poepsel S, Nogales E. Recent structural insights into Polycomb Repressive Complex 2 regulation and substrate binding. Biochemistry. 2019;58:346–54.
Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, Müller J. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat Struct Mol Biol. 2017;24:1039–47.
Laugesen A, Højfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell. 2019;74:8–18.
•• Deevy O, Bracken AP. PRC2 functions in development and congenital disorders. Development. 2019;146:dev181354–13.
•• Ezhkova E, Lien W-H, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–98.
•• Ferguson JW. Atit RP. A tale of two cities: the genetic mechanisms governing calvarial bone development genesis. 2018;129:e23246.
•• Schwarz D, Varum S, Zemke M, Scholer A, Baggiolini A, Draganova K, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141:867–77.
•• Ferguson J, Devarajan M, DiNuoscio G, Saiakhova A, Liu C-F, Lefebvre V, et al. PRC2 is dispensable in vivo for β-catenin-mediated repression of chondrogenesis in the mouse embryonic cranial mesenchyme. G3. 2018;8:491–503.
Snitow M, Lu M, Cheng L, Zhou S, Morrisey EE. Ezh2 restricts the smooth muscle lineage during mouse lung mesothelial development. Development. 2016;143:3733–41.
•• Ferguson JW, Devarajan M, Atit RP. Stage-specific roles of Ezh2 and retinoic acid signaling ensure calvarial bone lineage commitment. Dev Biol. 2018;443:173–87.
Ishii M, Sun J, Ting M-C, Maxson RE. The development of the calvarial bones and sutures and the pathophysiology of craniosynostosis. Curr Top Dev Biol Elsevier. 2015;115:131–56.
Fan X, A F Loebel D, Bildsoe H, E Wilkie E, P L tam P, Qin J, et al. Tissue interactions, cell signaling and transcriptional control in the cranial mesoderm during craniofacial development. AIMS Genetics 2016;3:74–98.
•• Dudakovic A, Camilleri E, Paradise CR, Samsonraj RM, Gluscevic M, Paggi CA, et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J Biol Chem. 2018;293:12894–907.
Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res American Society for Biochemistry and Molecular Biology. 2002;43:1773–808.
Carroll LS, Capecchi MR. Hoxc8 initiates an ectopic mammary program by regulating Fgf10 and Tbx3 expression and Wnt/β-catenin signaling. Development. 2015;142:4056–67.
Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, et al. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–81.
Funato N, Chapman SL, McKee MD, Funato H, Morris JA, Shelton JM, et al. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development (Cambridge, England). 2009;136:615–25.
Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201.
Lee Y, Lee J-Y, Kim MH. PI3K/Akt pathway regulates retinoic acid-induced Hox gene expression in F9 cells. Dev. Growth Differ. John Wiley & Sons, Ltd (10.1111); 2014;56:518–25.
Santagati F, Minoux M, Ren S-Y, Rijli FM. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development (Cambridge, England). 2005;132:4927–36.
Williams SS, Mear JP, Liang H-C, Potter SS, Aronow BJ, Colbert MC. Large-scale reprogramming of cranial neural crest gene expression by retinoic acid exposure. Physiol Genomics. 2004;19:184–97.
•• Mirzamohammadi F, Papaioannou G, Inloes JB, Rankin EB, Xie H, Schipani E, et al. Polycomb repressive complex 2 regulates skeletal growth by suppressing Wnt and TGF. Nat Commun. 2016;7:12047.
•• Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su I-H, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–35.
Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–30.
Han J, Ishii M, Bringas PJ, Maas RL, Maxson REJ, Chai Y. Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mechanisms of Development. 2007 ed. 2007;124:729–45.
•• Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the roles of Polycomb Repressive Complex 2 subunits in the control of skin development. J Invest Dermatol. 2016;136:1647–55.
Wang L, Jin Q, Lee J-E, Su I-H, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci. 2010;107:7317–22.
Zemke M, Draganova K, Klug A, Schöler A, Zurkirchen L, Gay MH-P, et al. Loss of Ezh2 promotes a midbrain-to-forebrain identity switch by direct gene derepression and Wnt-dependent regulation. BMC biology BioMed Central. 2015;13:103–14.
Yi SA, Han J, Han JW. Epigenetic role of nuclear S6K1 in early adipogenesis. BMB Rep. Korean Society for Biochemistry and Molecular Biology; 2016 Aug;:401–2.
Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol American Society for Microbiology Journals. 2007;27:5105–19.
Jung H-Y, Jun S, Lee M, Kim H-C, Wang X, Ji H, et al. PAF and EZH2 induce wnt/β-catenin signaling hyperactivation. Mol Cell Elsevier Inc. 2013;52:193–205.
•• Kumar D, Lassar AB. Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Reports The Authors. 2014;8:1419–31.
•• Hoffmeyer K, Junghans D, Kanzler B, Kemler R. Trimethylation and acetylation of b-catenin at lysine 49 represent key elements in ESC pluripotency. CellReports. ElsevierCompany; 2017;18:2815–2824.
Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175:1246–54.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–13.
Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56.
Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci. 2011;108:E149–58.
Rothberg JLM, Maganti HB, Jrade H, Porter CJ, Palidwor GA, Cafariello C, et al. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 2018;4:21–16.
Tee W-W, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–90.
Zhang Y, Liang J, Li Q. Coordinated regulation of retinoic acid signaling pathway by KDM5B and polycomb repressive complex 2. J. Cell. Biochem. John Wiley & Sons, Ltd; 2014;115:1528–38.
Kumar S, Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–7.
Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, et al. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–48.
Maclean G, Dollé P, Petkovich M. Genetic disruption of CYP26B1 severely affects development of neural crest derived head structures, but does not compromise hindbrain patterning. Dev Dyn. 2009;238:732–45.
Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–16.
•• Basu S, Mehreja R, Thiberge S, Chen M-T, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101:6355–60.
Funding
Funding is provided by NIH-NIDCR R01-DE18470 (RA), NIH-NIAMS T32 fellowship AR-007505 (JF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Timothy Nehila, James W. Ferguson, and Radhika P. Atit declare no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance
In order to efficiently and effectively apply orthopedic stem cell therapies, an in-depth understanding of specific genetic regulatory networks (GRN) in distinct musculoskeletal cell populations is essential. Epigenetic regulation is a “tool” that can be used to modulate regulation of GRN in order to alter cell differentiation programs. Manipulating epigenetic mechanisms has the potential to reveal new functions of GRN components that were not discernible by traditional loss- and gain-of-function approaches. Thus, the new knowledge gained from investigating epigenetic regulation of developing calvarial bone program and other tissues’ programs will inform tissue engineering and improve outcomes of genetic therapies in orthopedics.
This article is part of the Topical Collection on Skeletal Development
Rights and permissions
About this article
Cite this article
Nehila, T., Ferguson, J.W. & Atit, R.P. Polycomb Repressive Complex 2: a Dimmer Switch of Gene Regulation in Calvarial Bone Development. Curr Osteoporos Rep 18, 378–387 (2020). https://doi.org/10.1007/s11914-020-00603-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11914-020-00603-5