Abstract
Purpose of Review
While DDX41 mutation (m) is one of the most prevalent predisposition genes in adult myelodysplastic syndrome (MDS)/acute myeloid leukemia (AML), most patients do not always present with a family history of MDS/AML. In this review, we will be highlighting epidemiological data on DDX41m, roles of DDX41 in oncogenesis, mechanisms of clonal evolution with somatic DDX41m, and clinical phenotypes and management of MDS/AML in patients harboring DDX41m.
Recent Findings
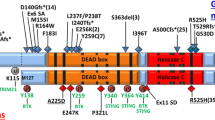
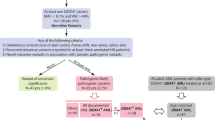
DDX41 encodes a DEAD-box helicase protein that is considered essential for cell growth and viability. High incidence of myeloid malignancies and other cancers in patients bearing DDX41m suggests that defects in DDX41 lead to loss of a tumor suppressor function, likely related to activities in RNA splicing and processing pathways. Seventy percent of cancer cases with DDX41m are associated with MDS/AML alone. More than 65% of familial cases harbor heterozygous germline frameshift mutations, of which p.D140Gfs*2 is the most common. A somatic DDX41m of the second allele is acquired in 70% of cases, leading to hematological malignancy. Myeloid neoplasms with DDX41m are typically characterized by long latency, high-risk disease at presentation with normal cytogenetics and without any additional molecular markers. Recent reports suggests that a subgroup of these patients have an indolent clinical course and have a better long-term survival compared to favorable or intermediate risk AML.
Summary
Distinct clinical/pathologic features and favorable outcomes in MDS/AML highlight the need for standardized classification and gene specific guidelines that could assist in management decisions in patients with DDX41m.

Similar content being viewed by others
References
Owen C, Barnett M, Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia—a review. Br J Haematol. 2008;140:123–32. https://doi.org/10.1111/j.1365-2141.2007.06909.x.
Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. https://doi.org/10.1182/blood-2016-03-643544.
Sébert M, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134:1441–4. https://doi.org/10.1182/blood.2019000909.
Maciejewski JP, Padgett RA, Brown AL, Müller-Tidow C. DDX41-related myeloid neoplasia. Semin Hematol. 2017;54:94–7. https://doi.org/10.1053/j.seminhematol.2017.04.007.
Omura H, et al. Structural and functional analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide. Sci Rep. 2016;6:34756. https://doi.org/10.1038/srep34756.
Polprasert C, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–70. https://doi.org/10.1016/j.ccell.2015.03.017.
Abou Dalle I, et al. Successful lenalidomide treatment in high risk myelodysplastic syndrome with germline DDX41 mutation. Am J Hematol. 2020;95:227–9. https://doi.org/10.1002/ajh.25610.
Wan Z, Han B. Clinical features of DDX41 mutation-related diseases: a systematic review with individual patient data. Ther Adv Hematol. 2021;12:20406207211032430. https://doi.org/10.1177/20406207211032433.
Alkhateeb HB, et al. Genetic features and clinical outcomes of patients with isolated and comutated DDX41-mutated myeloid neoplasms. Blood Adv. 2022;6:528–32. https://doi.org/10.1182/bloodadvances.2021005738.
Quesada AE, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol. 2019;94:757–66. https://doi.org/10.1002/ajh.25486.
Choi EJ, et al. Unique ethnic features of DDX41 mutations in patients with idiopathic cytopenia of undetermined significance, myelodysplastic syndrome, or acute myeloid leukemia. Haematologica. 2022;107:510–8. https://doi.org/10.3324/haematol.2020.270553.
Lewinsohn M, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–23. https://doi.org/10.1182/blood-2015-10-676098.
Ali MAM. DEAD-box RNA helicases: the driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021;296: 198352. https://doi.org/10.1016/j.virusres.2021.198352.
Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–65. https://doi.org/10.1038/ni.2091.
Parvatiyar K, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155. https://doi.org/10.1038/ni.2460.
Lee KG, et al. Bruton’s tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep. 2015;10:1055–65. https://doi.org/10.1016/j.celrep.2015.01.039.
Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe. 2015;17:478–88. https://doi.org/10.1016/j.chom.2015.02.021.
Stavrou, S., Aguilera, A. N., Blouch, K. & Ross, S. R. DDX41 recognizes RNA/DNA retroviral reverse transcripts and is critical for in vivo control of murine leukemia virus infection. mBio (2018) 9, https://doi.org/10.1128/mBio.00923-18.
Weinreb JT, et al. Excessive R-loops trigger an inflammatory cascade leading to increased HSPC production. Dev Cell. 2021;56:627-640.e625. https://doi.org/10.1016/j.devcel.2021.02.006.
Mosler T, et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat Commun. 2021;12:7314. https://doi.org/10.1038/s41467-021-27530-y.
Weinreb JT, Gupta V, Sharvit E, Weil R, Bowman TV. Ddx41 inhibition of DNA damage signaling permits erythroid progenitor expansion in zebrafish. Haematologica. 2022;107:644–54. https://doi.org/10.3324/haematol.2020.257246.
Polprasert C, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–70. https://doi.org/10.1016/j.ccell.2015.03.017.
Chlon TM, et al. Germline DDX41 mutations cause ineffective hematopoiesis and myelodysplasia. Cell Stem Cell. 2021. https://doi.org/10.1016/j.stem.2021.08.004.
Kadono M, et al. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol. 2016;44:745-754.e744. https://doi.org/10.1016/j.exphem.2016.04.017.
Wan Z, Han B. Clinical features of DDX41 mutation-related diseases: a systematic review with individual patient data. Therapeutic Advances in Hematology. 2021;12:20406207211032430. https://doi.org/10.1177/20406207211032433.
Choi EJ, et al. Unique ethnic features of DDX41 mutations in patients with idiopathic cytopenia of undetermined significance, myelodysplastic syndrome, or acute myeloid leukemia. Haematologica. 2021. https://doi.org/10.3324/haematol.2020.270553.
Polprasert C, et al. Novel DDX41 variants in Thai patients with myeloid neoplasms. Int J Hematol. 2020;111:241–6. https://doi.org/10.1007/s12185-019-02770-3.
Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. https://doi.org/10.1038/nature13035.
Chlon TM, et al. Germline DDX41 mutations cause ineffective hematopoiesis and myelodysplasia. Cell Stem Cell. 2021;28:1966-1981.e1966. https://doi.org/10.1016/j.stem.2021.08.004.
Kobayashi S, et al. Donor cell leukemia arising from preleukemic clones with a novel germline DDX41 mutation after allogenic hematopoietic stem cell transplantation. Leukemia. 2017;31:1020–2. https://doi.org/10.1038/leu.2017.44.
Gibson CJ, et al. Donor clonal hematopoiesis and recipient outcomes after transplantation. J Clin Oncol. 2021;40:189–201. https://doi.org/10.1200/JCO.21.02286.
Berger G, et al. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2016;31:520. https://doi.org/10.1038/leu.2016.310.
Li P, et al. AML with germline DDX41 variants is a clinicopathologically distinct entity with an indolent clinical course and favorable outcome. Leukemia. 2021. https://doi.org/10.1038/s41375-021-01404-0.
Alkhateeb HB, et al. Genetic features and clinical outcomes of patients with isolated and comutated DDX41-mutated myeloid neoplasms. Blood Adv. 2021. https://doi.org/10.1182/bloodadvances.2021005738.
Negoro E, et al. Molecular predictors of response in patients with myeloid neoplasms treated with lenalidomide. Leukemia. 2016;30:2405–9. https://doi.org/10.1038/leu.2016.228.
Madanat YF, et al. Genomic biomarkers predict response/resistance to lenalidomide in non-Del(5q) myelodysplastic syndromes. Blood. 2018;132:1797. https://doi.org/10.1182/blood-2018-99-114681.
Li R, Sobreira N, Witmer PD, Pratz KW, Braunstein EM. Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Haematologica. 2016;101:e228-231. https://doi.org/10.3324/haematol.2015.139790.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Human and Animal Rights and Informed Consent.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Germline Predisposition to Myeloid Neoplasms.
Rights and permissions
About this article
Cite this article
Badar, T., Chlon, T. Germline and Somatic Defects in DDX41 and its Impact on Myeloid Neoplasms. Curr Hematol Malig Rep 17, 113–120 (2022). https://doi.org/10.1007/s11899-022-00667-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-022-00667-3