Abstract
Purpose
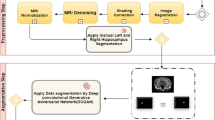
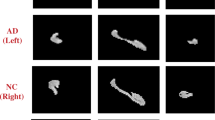
Knowing the course of Alzheimer’s disease is very important to prevent the deterioration of the disease, and accurate segmentation of sensitive lesions can provide a visual basis for the diagnosis results. This study proposes an improved end-to-end dual-functional 3D convolutional neural network for segmenting bilateral hippocampi from 3D brain MRI scans and diagnosing AD progression states simultaneously.
Methods
The proposed neural network is based on the V-Net and adopts an end-to-end structure. In order to relieve the excessive amount of convolutional parameters at the bottom of the V-Net, we change them to bottleneck architecture. Based on the segmentation network, we establish a classification network for diagnosing pathological states of brain. In order to balance the two tasks of hippocampi segmentation and brain pathological states diagnosis, we designed a unique loss function. This study included 132 samples, of which 100 were selected as training, and the remaining 32 were used to test the performance of our model. During training, we adopted fivefold cross-validation method.
Results
We selected the intersection over union and dice coefficient to evaluate the hippocampus segmentation performance, while the brain pathological states diagnosis performance was evaluated by accuracy, specificity, sensitivity, precision and F1 score. By using the proposed neural network, the left hippocampi segmentation Iou and dice coefficient reach 0.8240 ± 0.020 and 0.9035 ± 0.020, respectively. The right hippocampi Iou and dice coefficient reach 0.8454 ± 0.023 and 0.9162 ± 0.023, respectively. The accuracy, specificity, sensitivity, precision and F1 score of three-category classification of brain pathology are 84%, 92%, 84%, 86% and 85%, respectively.
Conclusion
The proposed neural network has two functions of brain pathological states diagnosis and bilateral hippocampi segmentation with higher robustness and accuracy, respectively. The segmented bilateral hippocampi can be used as a reference for clinical decision making or intervention.









Similar content being viewed by others
References
Sami S, Williams N, Hughes LE, Cope TE, Rittman T, Coyle-Gilchrist ITS, Henson RN, Rowe JB (2018) Neurophysiological signatures of Alzheimer’s disease and frontotemporal lobar degeneration: pathology versus phenotype. Brain 141(8):2500–2510
Zhang Y, Wang S, Phillips P, Dong Z, Ji G, Yang J (2015) Detection of Alzheimer’s disease and mild cognitive impairment based on structural volumetric MR images using 3D-DWT and WTA-KSVM trained by PSOTVAC. Biomed Signal Process Control 21:58–73
Cassani R, Estarellas M, San-Martin R, Fraga FJ, Falk TH (2018) Systematic review on resting-state EEG for Alzheimer’s disease diagnosis and progression assessment. Dis Markers 2018:1–26
Wolk DA, Das SR, Mueller SG, Weiner MW, Yushkevich PA (2017) Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer’s disease. Neurobiol Aging 49:204–213
Elder GJ, Mactier K, Colloby SJ, Watson R, Blamire AM, O’Brien JT, Taylor JP (2017) The influence of hippocampal atrophy on the cognitive phenotype of dementia with Lewy bodies. Int J Geriatr Psychiatry 32(11):1182–1189
Wang L, Pavlidis T (1993) Direct gray-scale extraction of features for character recognition. IEEE Trans Pattern Anal Mach Intell 15(10):1053–1067
Ma WY, Manjunath BS (1996) Texture features and learning similarity. In: IEEE computer society conference on computer vision and pattern recognition, vol 1, number 1, pp 425–430
Amit Y, Geman D, Wilder K (1997) Joint induction of shape features and tree classifiers. IEEE Trans Pattern Anal Mach Intell 19(11):1300–1305
Zhang S, Sun F, Wang N, Zhang C, Yu Q, Zhang M, Babyn P, Zhong H (2019) Computer-aided diagnosis (CAD) of pulmonary nodule of thoracic CT image using transfer learning. J Digit Imaging 2019:1–13
Zhu HC, Cheng HW, Yang XS, Fan Y (2016) Metric learning for multi-atlas based segmentation of hippocampus. Neuroinformatics 15(1):1–10
Jiang X, Zhou Z, Ding X, Deng X, Zou L, Li B (2017) Level set based hippocampus segmentation in MR images with improved initialization using region growing. Comput Math Methods Med 1:1–11
Sch C, Laptev I, Caputo B (2004) Recognizing human actions: a local SVM approach. In: International conference on pattern recognition
Beheshti I, Maikusa N, Matsuda H, Demirel H, Anbarjafari G (2017) Histogram-based feature extraction from individual gray matter similarity-matrix for Alzheimer’s disease classification. J Alzheimer’s Dis (JAD) 55(4):1571
Altaf T, Anwar SM, Gul N, Majeed MN, Majid M (2018) Multi-class Alzheimer’s disease classification using image and clinical features. Biomed Signal Process Control 43:64–74
Chen Y, Shi B, Wang Z, Zhang P (2017) Hippocampus segmentation through multi-view ensemble ConvNets. In: Int Symp Biomed Imaging
Xie Z, Gillies D (2018) Near real-time hippocampus segmentation using patch-based canonical neural network. https://arxiv.org/pdf/1807.05482.pdf
Liu J, Li M, Lan W, Wu FX, Pan Y, Wang J (2018) Classification of Alzheimer’s disease using whole brain hierarchical network. IEEE/ACM Trans Comput Biol Bioinf 15(2):624–632
Sarraf S, Tofighi G (2016) Classification of Alzheimer’s disease structural MRI data by deep learning convolutional neural networks. https://arxiv.org/abs/1603.08631
Milletari F, Navab N, Ahmadi SA (2016) V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: International conference on 3D vision
Sherman R (2018) A volumetric convolutional neural network for brain tumor segmentation. https://arxiv.org/pdf/1811.02654.pdf
Xu L, Tetteh G, Lipkova J, Zhao Y, Li H, Christ P, Piraud M, Buck A, Shi K, Menze BH (2018) Automated whole-body bone lesion detection for multiple myeloma on 68 Ga-pentixafor PET/CT imaging using deep learning methods. Contrast Media Mol Imaging 2018:1–11
Farahnak-Ghazani F, Baghshah MS (2016) Multi-label classification with feature-aware implicit encoding and generalized cross-entropy loss. In: Iranian conference on electrical engineering
Ketkar N (2017) Introduction to PyTorch. In: Deep Learning with Python. Apress, Berkeley, CA, pp 195–208
Bock S, Goppold J, Weiß M (2018) An improvement of the convergence proof of the adam-optimizer. https://arxiv.org/pdf/1804.10587.pdf
Chen Y, Shi B, Wang Z, Sun T, Smith CD, Liu J (2017) Accurate and consistent hippocampus segmentation through convolutional LSTM and view ensemble. Mach Learn Med Imaging. https://doi.org/10.1007/978-3-319-67389-9_11
Rieke J, Eitel F, Weygandt M, Haynes JD (2018) Visualizing convolutional networks for MRI-based diagnosis of Alzheimer’s disease. https://arxiv.org/pdf/1808.02874.pdf
Korolev S, Safiullin A, Belyaev M, Dodonova Y (2017) Residual and plain convolutional neural networks for 3D brain MRI classification. In: International symposium on biomedical imaging, vol 2017, pp 835–838
Dolph C, Alam M, Shboul Z, Samad M (2017) Deep learning of texture and structural features for multiclass Alzheimer’s disease classification. In: International joint conference on neural networks
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain patient data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, J., Yan, S., Song, C. et al. Dual-functional neural network for bilateral hippocampi segmentation and diagnosis of Alzheimer’s disease. Int J CARS 15, 445–455 (2020). https://doi.org/10.1007/s11548-019-02106-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-02106-w