Abstract
Purpose
To develop a CT texture-based model able to predict epidermal growth factor receptor (EGFR)-mutated, anaplastic lymphoma kinase (ALK)-rearranged lung adenocarcinomas and distinguish them from wild-type tumors on pre-treatment CT scans.
Materials and methods
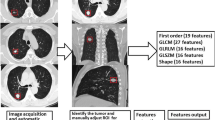
Texture analysis was performed using proprietary software TexRAD (TexRAD Ltd, Cambridge, UK) on pre-treatment contrast-enhanced CT scans of 84 patients with metastatic primary lung adenocarcinoma. Textural features were quantified using the filtration-histogram approach with different spatial scale filters on a single 5-mm-thick central slice considered representative of the whole tumor. In order to deal with class imbalance regarding mutational status percentages in our population, the dataset was optimized using the synthetic minority over-sampling technique (SMOTE) and correlations with textural features were investigated using a generalized boosted regression model (GBM) with a nested cross-validation approach (performance averaged over 1000 resampling episodes).
Results
ALK rearrangements, EGFR mutations and wild-type tumors were observed in 19, 28 and 37 patients, respectively, in the original dataset. The balanced dataset was composed of 171 observations. Among the 29 original texture variables, 17 were employed for model building. Skewness on unfiltered images and on fine texture was the most important features. EGFR-mutated tumors showed the highest skewness while ALK-rearranged tumors had the lowest values with wild-type tumors showing intermediate values. The average accuracy of the model calculated on the independent nested validation set was 81.76% (95% CI 81.45–82.06).
Conclusion
Texture analysis, in particular skewness values, could be promising for noninvasive characterization of lung adenocarcinoma with respect to EGFR and ALK mutations.







Similar content being viewed by others
Abbreviations
- ALK:
-
Anaplastic lymphoma kinase
- CT:
-
Computed tomography
- CTTA:
-
CT texture analysis
- EGFR:
-
Epidermal growth factor receptor
- FBP:
-
Filtered back-projection
- FFPE:
-
Formalin-fixed paraffin-embedded
- FISH:
-
Fluorescence in situ hybridization
- FOV:
-
Field of view
- GBM:
-
Generalized boosted regression model
- KV:
-
Kilovolt
- MAS:
-
Milliampere per seconds
- ML:
-
Milliliters
- MRMR:
-
Minimum redundancy maximal relevancy filter
- NSCLC:
-
Non-small cell lung cancer
- S:
-
Seconds
- SD:
-
Standard deviation
- MOTE:
-
Synthetic minority oversampling technique
- SSF:
-
Spatial scale filter
References
Fitzmaurice C, Allen C et al (2017) Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol 3(524):548. https://doi.org/10.1001/jamaoncol.2016.5688
Barlesi F, Mazieres J, Merlio J-P et al (2016) Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the french cooperative thoracic intergroup (IFCT). The Lancet 387:1415–1426. https://doi.org/10.1016/S0140-6736(16)00004-0
Hanna N, Johnson D, Temin S et al (2017) Systemic therapy for stage IV non–small-cell lung cancer: American society of clinical oncology clinical practice guideline update. JCO 35:3484–3515. https://doi.org/10.1200/JCO.2017.74.6065
Novello S, Barlesi F, Califano R et al (2016) Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 27:v1–v27. https://doi.org/10.1093/annonc/mdw326
Rosell R, Carcereny E, Gervais R et al (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13:239–246. https://doi.org/10.1016/S1470-2045(11)70393-X
Kwak EL, Bang Y-J, Camidge DR et al (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363:1693–1703. https://doi.org/10.1056/NEJMoa1006448
Choi YR, An JY, Kim MK et al (2013) The diagnostic efficacy and safety of endobronchial ultrasound-guided transbronchial needle aspiration as an initial diagnostic tool. Korean J Intern Med 28:660. https://doi.org/10.3904/kjim.2013.28.6.660
Lian W, Ouyang Y (2017) CT-guided aspiration lung biopsy for EGFR and ALK gene mutation analysis of lung cancer. Oncology Letters 13:3415–3422. https://doi.org/10.3892/ol.2017.5921
(2018) Clinical implications of transbronchial biopsy for surgically-resected non-small cell lung cancer. In Vivo https://doi.org/10.21873/invivo.11295
Hofman P (2017) ALK status assessment with liquid biopsies of lung cancer patients. Cancers 9:106. https://doi.org/10.3390/cancers9080106
Lin C-C, Huang W-L, Wei F et al (2015) Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev Mol Diagn 15:1427–1440. https://doi.org/10.1586/14737159.2015.1094379
Liu Y, Kim J, Balagurunathan Y et al (2016) Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clinical Lung Cancer 17:441-448.e6. https://doi.org/10.1016/j.cllc.2016.02.001
Nair JKR, Saeed UA, McDougall CC et al (2020) Radiogenomic models using machine learning techniques to predict EGFR mutations in non-small cell lung cancer. Can Assoc Radiol J. https://doi.org/10.1177/0846537119899526
Mei D, Luo Y, Wang Y, Gong J (2018) CT texture analysis of lung adenocarcinoma: can radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 18:52. https://doi.org/10.1186/s40644-018-0184-2
Weiss GJ, Ganeshan B, Miles KA et al (2014) Noninvasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS ONE 9:e100244. https://doi.org/10.1371/journal.pone.0100244
Digumarthy SR, Padole AM, Gullo RL et al (2019) Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status. Medicine 98 https://doi.org/10.1097/MD.0000000000013963
Yoon HJ, Sohn I, Cho JH, et al (2015) Decoding Tumor Phenotypes for ALK ROS1 and RET Fusions in Lung Adenocarcinoma Using a Radiomics Approach, Medicine 94: 1753 https://doi.org/10.1097/MD.0000000000001753
Martin V, Bernasconi B, Merlo E et al (2015) ALK testing in lung adenocarcinoma: technical aspects to improve FISH evaluation in daily practice. J Thorac Oncol 10:595–602. https://doi.org/10.1097/JTO.0000000000000444
Ravanelli M, Agazzi GM, Ganeshan B et al (2018) CT texture analysis as predictive factor in metastatic lung adenocarcinoma treated with tyrosine kinase inhibitors (TKIs). Eur J Radiol 109:130–135. https://doi.org/10.1016/j.ejrad.2018.10.016
Miles KA, Ganeshan B, Hayball MP (2013) CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging 13:400–406. https://doi.org/10.1102/1470-7330.2013.9045
Ng F, Kozarski R, Ganeshan B, Goh V (2013) Assessment of tumor heterogeneity by CT texture analysis: can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur J Radiol 82:342–348. https://doi.org/10.1016/j.ejrad.2012.10.023
Shafiq-ul-Hassan M, Zhang GG, Latifi K et al (2017) Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 44:1050–1062. https://doi.org/10.1002/mp.12123
Blagus R, Lusa L (2013) SMOTE for high-dimensional class-imbalanced data. BMC Bioinformatics 14:106. https://doi.org/10.1186/1471-2105-14-106
Peng H, Long F, Ding C (2005) Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27:1226–1238. https://doi.org/10.1109/TPAMI.2005.159
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Lin JJ, Riely GJ, Shaw AT (2017) Targeting ALK: precision medicine takes on drug resistance. Cancer Discov 7:137–155. https://doi.org/10.1158/2159-8290.CD-16-1123
Cai W, Lin D, Wu C et al (2015) Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol 33:3701–3709. https://doi.org/10.1200/JCO.2014.58.8293
Zito Marino F, Marino FZ, Liguori G et al (2015) Intratumor heterogeneity of ALK-rearrangements and homogeneity of EGFR-mutations in mixed lung adenocarcinoma. PLoS ONE 10:e0139264. https://doi.org/10.1371/journal.pone.0139264
Kobayashi Y, Mitsudomi T (2016) Not all epidermal growth factor receptor mutations in lung cancer are created equal: perspectives for individualized treatment strategy. Cancer Sci 107:1179–1186. https://doi.org/10.1111/cas.12996
Nagahashi M, Sato S, Yuza K et al (2018) Common driver mutations and smoking history affect tumor mutation burden in lung adenocarcinoma. J Surg Res 230:181–185. https://doi.org/10.1016/j.jss.2018.07.007
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Because of the retrospective nature of this study, the requirement for informed consent was waived by the local institutional review board of University of Brescia.
Ethical approval
Ethical approval was waived by the local institutional review board of University of Brescia (NP 4424) in view of the retrospective nature of the study and all the procedures being performed were part of the routine care. The study was conducted in accordance with the ethical standards of the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Agazzi, G.M., Ravanelli, M., Roca, E. et al. CT texture analysis for prediction of EGFR mutational status and ALK rearrangement in patients with non-small cell lung cancer. Radiol med 126, 786–794 (2021). https://doi.org/10.1007/s11547-020-01323-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-020-01323-7