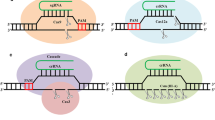
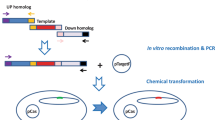
Abstract
The innovative CRISPR–Cas based genome editing technology provides some functionality and advantages such as the high efficiency and specificity as well as ease of handling. Both aspects of the CRISPR–Cas9 system including genetic engineering and gene regulation are advantageously applicable to the construction of microbial cell factories. As one of the most extensively used cell factories, E. coli has been engineered to produce various high value-added chemical compounds such as pharmaceuticals, biochemicals, and biofuels. Therefore, to improve the production of valuable metabolites, many investigations have been performed by focusing on CRISPR–Cas- based metabolic engineering of this host. In the current review, the biology underlying CRISPR–Cas9 system was briefly explained and then the applications of CRISPR–Cas9/CRISPRi tools were considered for cell factory construction in E. coli.




Similar content being viewed by others
References
Abdelaal AS, Yazdani SS. (2020). Development and use of CRISPR in industrial applications. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 177–197
Adiego-Pérez B, Randazzo P, Daran JM, Verwaal R, Roubos JA, Daran-Lapujade P, Van Der Oost J. (2019). Multiplex genome editing of microorganisms using CRISPR-Cas FEMS microbiology letters 366:fnz086
Alkhnbashi OS, Meier T, Mitrofanov A, Backofen R, Voß B (2020) CRISPR-Cas Bioinformatics Methods 172:3–11
An J, Zhang W, Jing X, Nie Y, Xu Y. (2020). Reconstitution of TCA cycle involving l-isoleucine dioxygenase for hydroxylation of l-isoleucine in Escherichia coli using CRISPR-Cas9 3 Biotech 10:1–10
Bae S, Park J, Kim J-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases Bioinformatics 30:1473–1475
Bhushan K. (2020). Evolution and molecular mechanism of CRISPR/Cas9 systems. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 15–25
Boch J et al (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512
Bortesi L, Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology advances, 33: 41–52
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Chaikind B, Van Rossum HM, Miller A, Perkovich P, Szyjka S, Patel K. (2020). Applications of CRISPRI in high throughput metabolic engineering. Google Patents
Charpentier E, Elsholz A, Marchfelder A. (2019). CRISPR-Cas: more than ten years and still full of mysteries. Taylor & Francis
Chen S, Yao Y, Zhang Y, Fan G. (2020). CRISPR system: Discovery, development and off-target detection Cellular Signalling, 109577
Cho S, Choe D, Lee E, Kim SC, Palsson B, Cho B-K. (2018). High-level dCas9 expression induces abnormal cell morphology in Escherichia coli ACS synthetic biology 7:1085–1094
Cho S, Shin J, Cho B-K (2018) Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci 19:1089
Choudhary M, Joshi S, Singh P, Srivastava N. (2020). Biofuel production from lignocellulosic biomass: Introduction and metabolic engineering for fermentation scale-up. In: Genetic and Metabolic Engineering for Improved Biofuel Production from Lignocellulosic Biomass. Elsevier, pp 1–12
Chuai G-h, Wang Q-L, Liu Q (2017) In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol 35:12–21
Cress BF, Leitz QD, Kim DC, Amore TD, Suzuki JY, Linhardt RJ, Koffas MA. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production Microbial cell factories 16:10
Cress BF et al. (2015). CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Syn Biol 4:987–1000
Das M, Patra P, Ghosh A (2020) Metabolic engineering for enhancing microbial biosynthesis of advanced biofuels. Renew Sustain Energy Rev 119:109562
Dasgupta A, Chowdhury N, De RK (2020) Metabolic pathway engineering: perspectives and applications. Comput Methods Programs Biomed 192:105436
Deyell M, Ameta S, Nghe P. (2019). Large scale control and programming of gene expression using CRISPR. In: Seminars in cell & developmental biology, Elsevier
Ding W, Weng H, Du G, Chen J, Kang Z. (2017). 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli. J Indus Microbiol Biotechnol 44:1127–1135
Doench JG et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 34:184
Dong C. (2019). Programming bacterial gene expression using synthetic CRISPR-Cas transcriptional regulators.
Dong X et al (2020) CRISPRi-guided multiplexed fine-tuning of metabolic flux for enhanced Lacto-N-neotetraose production in Bacillus subtilis. J Agricul Food Chem 68:2477–2484
Ebrahimi V, Hashemi A. (2020). Challenges of in vitro genome editing with CRISPR/Cas9 and possible solutions: a review Gene, 144813
Fokum E et al. (2019). Metabolic engineering of bacterial strains using CRISPR/Cas9 systems for biosynthesis of value-added products. Food Biosci, 28:125–132
Fujiwara R, Noda S, Tanaka T, Kondo A. (2020). Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose–xylose co-substrate Nature Communications, 11:1–12
Geng Y, Deng Z, Sun Y. (2016). An insight into the protospacer adjacent motif of Streptococcus pyogenes Cas9 with artificially stimulated RNA-guided-Cas9 DNA cleavage flexibility. RSC Adv 6:33514–33522
Guo T, Xin Y, Zhang Y, Gu X, Kong J (2019) A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb Cell Fact 18:22
Hashemi A (2018) CRISPR-Cas system as a genome engineering platform: applications in biomedicine and biotechnology. Curr Gene Ther 18:115–124
Heo M-J, Jung H-M, Um J, Lee S-W, Oh M-K. (2017). Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli. ACS Synth Biol 6:182–189
Herai RH. (2019). Avoiding the off-target effects of CRISPR/cas9 system is still a challenging accomplishment for genetic transformation. Gene 700:176–178
Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259
Hsu PD et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol, 31:827
Huminiecki L, Horbańczuk J (2018) The functional genomic studies of resveratrol in respect to its anti-cancer effects. Biotechnol Adv 36:1699–1708
Ibrahim A, ÖZSÖZ M SZ, Tirah G, Gideon O (2019) Genome engineering using the CRISPR Cas9 system. J Biomed Pharm Sci, 2:2
Kim SK, Han GH, Seong W, Kim H, Kim S-W, Lee D-H, Lee S-G (2016) CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab Eng 38:228–240
Kim SK, Seong W, Han GH, Lee D-H, Lee S-G (2017) CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Fact 16:188
Koonin EV, Makarova KS (2019) Origins and evolution of CRISPR-Cas systems. Philosoph Trans Royal Soc B 374:20180087
Li D, Zhou H, Zeng X. (2019). Battling CRISPR-Cas9 off-target genome editing. Springer
Li Q-S, Li Y, Deora GS, Ruan B-F (2019) Derivatives and analogues of resveratrol: recent advances in structural modification. Mini Rev Med Chem 19:809–825
Li Q, Seys FM, Minton NP, Yang J, Jiang Y, Jiang W, Yang S. (2019) CRISPR–Cas9D10A nickase‐assisted base editing in the solvent producer Clostridium beijerinckii. Biotechnol Bioeng, 116:1475–1483
Li S, Jendresen CB, Grünberger A, Ronda C, Jensen SI, Noack S, Nielsen AT (2016) Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab Eng 38:274–284
Li Y et al. (2015). Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metabolic Eng, 31:13–21
Liang L, Liu R, Garst AD, Lee T, Beckham GT, Gill RT (2017) CRISPR EnAbled trackable genome engineering for isopropanol production in Escherichia coli. Metab Eng 41:1–10
Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. (2015). CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics, 31:3676–3678
Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W (2019) Methodologies for improving HDR efficiency. Front Genet 9:691
Lunge A, Choudhary E, Sharma R, Gupta R, Agarwal N. (2020). Functional understanding of CRISPR interference: its advantages and limitations for gene silencing in bacteria. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 199–218
Lv L, Ren Y-L, Chen J-C, Wu Q, Chen G-Q. (2015). Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P (3HB-co-4HB) biosynthesis. Metabol Eng, 29:160–168
Ma M, Ye AY, Zheng W, Kong L. (2013). A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. BioMed Res Int 2013
Macpherson CR, Scherf A (2015) Flexible guide-RNA design for CRISPR applications using protospacer workbench. Nat Biotechnol 33:805
Makarova KS et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol, 13:722–736
Marcellin E, Nielsen LK (2018) Advances in analytical tools for high throughput strain engineering. Curr Opin Biotechnol 54:33–40
McCarty NS, Graham AE, Studená L, Ledesma-Amaro R (2020) Multiplexed CRISPR technologies for gene editing and transcriptional regulation Nature. Communications 11:1–13
Mitsui R, Yamada R, Ogino H (2019) CRISPR system in the yeast Saccharomyces cerevisiae and its application in the bioproduction of useful chemicals. World J Microbiol Biotechnol 35:111
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res, 42:W401-W407
Moon SB, Ko J-H, Kim Y-S (2019) Recent advances in the CRISPR genome editing tool set. Exp Mol Med 51:1–11
Mougiakos I, Bosma EF, Ganguly J, van der Oost J, van Kranenburg R (2018) Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: advances and prospects. Curr Opin Biotechnol 50:146–157
Naito Y, Hino K, Bono H, Ui-Tei K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics, 31:1120–1123
Nandy D, Maity A, Mitra AK (2020) Target-specific gene delivery in plant systems and their expression: Insights into recent developments. J Biosci 45:30
Nishimasu H et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 156:935–949
O’Brien A, Bailey TL. (2014). GT-Scan: identifying unique genomic targets Bioinformatics 30:2673–2675
Pan S, Reed JL (2018) Advances in gap-filling genome-scale metabolic models and model-driven experiments lead to novel metabolic discoveries. Curr Opin Biotechnol 51:103–108
Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ (2018) Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol 13:389–396
Prykhozhij SV, Vinothkumar Rajan DG, Berman JN. (2015). CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences, PloS one, 10
Reisch CR, Prather KL. (2015). The no-SCAR (S carless C as9 A ssisted R ecombineering) system for genome editing in Escherichia coli. Sci Rep, 5:1–12
Satowa D et al. (2020). Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl‐CoA supply. Biotechnol Bioeng
Schultenkämper K, Brito LF, Wendisch VF. (2020). Impact of CRISPR interference on strain development in biotechnology. Biotechnol Appl Biochem
Shen C-C, Hsu M-N, Chang C-W, Lin M-W, Hwu J-R, Tu Y, Hu Y-C. (2019). Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res, 47:e13-e13
Shmakov S et al. (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems, Mol cell, 60:385–397
Singh V. (2020). An introduction to genome editing CRISPR-Cas systems. In: Genome Engineering via CRISPR-Cas9 System. Elsevier, pp 1–13
Sledzinski P, Nowaczyk M, Olejniczak M (2020) Computational tools and resources supporting CRISPR-Cas. Exp Cell 9:1288
Song M. (2017). The CRISPR/Cas9 system: Their delivery, in vivo and ex vivo applications and clinical development by startups. Biotechnol Progress, 33:1035–1045
Song X, Huang H, Xiong Z, Ai L, Yang S. (2017). CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl Environ Microbiol, 83:e01259–01217
Standage-Beier K, Zhang Q, Wang X (2015) Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS Synth Biol 4:1217–1225
Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool, PloS one, 10
Terns RM, Terns MP (2014) CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet 30:111–118
Tian J, Yang G, Gu Y, Sun X, Lu Y, Jiang W. (2020). Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in industrial Streptomyces bioRxiv
Tian P, Wang J, Shen X, Rey JF, Yuan Q, Yan Y (2017) Fundamental CRISPR-Cas9 tools and current applications in microbial systems. Synth Sys Biotechnolo 2:219–225
Trimidal SG et al (2019) Can designer indels be tailored by gene editing? can indels be customized? BioEssays 41:1900126
Van Der Oost J, Westra ER, Jackson RN, Wiedenheft B (2014) Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol 12:479–492
Vento JM, Crook N, Beisel CL (2019) Barriers to genome editing with CRISPR in bacteria. J Indus Microbiol Biotechnol 46:1327–1341
Vigouroux A, Bikard D. (2020). CRISPR tools to control gene expression in bacteria. Microbiol Mol Biol Rev, 84
Wang B, Guo Y, Xu Z, Tu R, Wang Q. (2020). Genomic, transcriptomic, and metabolic characterizations of Escherichia coli adapted to branched-chain higher alcohol tolerance. Appl Microbiol Biotechnol, 1–14
Wang Q, Zeng W, Zhou J (2019) Effect of gene knock-out of L-tyrosine transport system on L-tyrosine production in Escherichia coli Sheng wu gong cheng xue bao. Chin J Biotechnol 35:1247–1255
Wilson LO, O’Brien AR, Bauer DC (2018) The current state and future of CRISPR-Cas9 gRNA design tools. Front Pharmacol 9:749
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 16:218
Wright AV, Nuñez JK, Doudna JA (2016) Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164:29–44
Wu J, Du G, Chen J, Zhou J (2015) Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci Rep 5:13477
Wu J, Zhang X, Zhu Y, Tan Q, He J, Dong M (2017) Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci Rep 7:1–15
Wu J, Zhou P, Zhang X, Dong M (2017) efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J Indus Microbiol Biotechnol 44:1083–1095
Wu M-Y, Sung L-Y, Li H, Huang C-H, Hu Y-C (2017) Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1, 4-BDO biosynthesis. ACS Synth Biol 6:2350–2361
Wu Y et al (2020) Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Res 48:996–1009
Xia J et al (2016) Expression of Shewanella frigidimarina fatty acid metabolic genes in E. coli by CRISPR/cas9-coupled lambda Red recombineering. Biotechnol let 38:117–122
Xie S, Shen B, Zhang C, Huang X, Zhang Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS one, 9
Xu H et al (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res 25:1147–1157
Xu S, Wang Q, Zeng W, Li Y, Shi G, Zhou J. (2020). Construction of a heat-inducible Escherichia coli strain for efficient de novo biosynthesis of L-tyrosine. Process Biochem
Yin H et al (2018) Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chemical Biol 14:311
Zhan T, Rindtorff N, Betge J. (2019). Ebert MP, Boutros M CRISPR/Cas9 for cancer research and therapy. In: Seminars in cancer biology, Elsevier, pp 106–119
Zhang B, Zhang X-M, Wang W, Liu Z-Q, Zheng Y-G (2019) Metabolic engineering of Escherichia coli for d-pantothenic acid production. Food Chem 294:267–275
Zhang H, Cheng Q-X, Liu A-M, Zhao G-P, Wang J (2017) A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol 8:812
Zhu LJ, Holmes BR, Aronin N, Brodsky MH. (2014). CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems, PloS one, 9
Ziegler M, Takors R (2020) Reduced and minimal cell factories in bioprocesses: towards a streamlined chassis. Minimal Cells: Design. Construction, Biotechnological Applications. Springer, pp 1–44
Zou X et al (2020) Pathway construction and metabolic engineering for fermentative production of β-alanine in Escherichia coli. Appl Microbiol Biotechnol 104:2545–2559
Acknowledgements
This work was supported by the research deputy of Shahid Beheshti University of Medical Sciences in Tehran, Iran.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hashemi, A. CRISPR–Cas9/CRISPRi tools for cell factory construction in E. coli. World J Microbiol Biotechnol 36, 96 (2020). https://doi.org/10.1007/s11274-020-02872-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-020-02872-9