Abstract
Is the origin of gibbon ape leukemia virus (GALV) human after all? When GALV was discovered and found to cause neoplastic disease in gibbons, it stimulated a great deal of research including investigations into the origins of this virus. A number of publications have suggested that the GALV progenitor was a retrovirus present in one of several species of South East Asian rodents that had close contact with captive gibbons. However, there are no published retroviral sequences from any South East Asian species to support this view. Here we present an alternative hypothesis that the origin of GALV is a virus closely related to Melomys burtoni retrovirus, and that this virus infected human patients in Papua New Guinea from whom biological material was obtained or in some way contaminated these samples. This material we propose contained infectious MbRV-related virus that was then unwittingly introduced into gibbons which subsequently developed GALV infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibbon ape leukemia virus (GALV) was first isolated in captive gibbons housed at the US Army-SEATO medical research facility in Bangkok, Thailand in the late 1960s. This novel retrovirus is oncogenic and causes a range of lymphoid tumours in sub-human primates. Gibbon ape leukemia virus is related to murine leukemia viruses, and a number of gamma retroviruses from South East Asian rodents have been proposed to be the origin of GALV. However, there are no published proviral sequences from any of these rodent hosts which share a sufficiently high degree of identity to GALV to support this hypothesis. The closest GALV-related proviral sequences are those of Melomys burtoni retrovirus (MbRV), isolated from the grassland melomys, a rodent restricted to north eastern Australia and Papua New Guinea (PNG) and MelWMV isolated from a Melomys burtoni sub species from islands in Wallacia. A careful review the SEATO facility’s reports shows that gibbons were injected with biological material from human patients from PNG up to a year or more before the first GALV strain (GALV-SEATO) was identified. A second strain of GALV (GALV-BR) was isolated independently from gibbons housed at a US research facility that had no contact with GALV-infected gibbons but which had been injected with biological material derived from gibbons injected with human brain samples also derived from PNG. There is a high degree of sequence identity shared by GALV and MbRV, the MbRV host is widespread in Papua New Guinea and two strains of GALV have been isolated from gibbons following the injection of material of human origin derived from Papua New Guinea. Together these observations lend weight to the alternative hypothesis that MbRV has made a species jump from rodents to humans in Papua New Guinea and that MbRV contaminated human-derived material unwittingly injected into gibbons was the origin of GALV.
Background
The US Army-SEATO medical research facility was established in 1960 in Bangkok, Thailand (http://www.afrims.org/afrims-history.html). Gibbons were first housed at the facility in 1965/6 (http://www.afrims.org/weblib/eapr/1966/APR66p147-152.pdf) and were used primarily for research on Dengue Fever and malaria. In 1971, the facility notified the first case of malignant lymphoma in a white-handed gibbon (Hylobates lar) [1]. Prior to this, there were few reports of lymphoid tumours in non-human primates and only two in gibbons [2, 3]. In 1972, Kawakami et al. described an association between gammaretroviruses (C-type virus) and lymphosarcoma in gibbons [4]. This virus has been named Gibbon ape leukemia virus (GALV) [5]. It appears the first known cases of GALV infection and concomitant lymphosarcoma occurred at the SEATO facility. There are no reports of GALV infection in wild gibbons.
These observations raise strong suspicion that prior to the 1960s, gibbons were naïve to this virus and that GALV exposure, infection and clinical expression were idiosyncratic to the captive environment of the SEATO facility. This assessment that SEATO gibbons were simply an opportunistic host for GALV is indirectly supported by a recent survey of gibbons in zoological institutions in North America that described serological evidence of exposure to GALV-like antigens in gibbons but failed to detect evidence of intact virus or evidence of clinical association [6]. Overall, there was no compelling corroboration from these records that would support gibbons as a reservoir host for GALV. Unfortunately at the moment, there is no information on the GALV status of free-ranging gibbon populations that could help rule out gibbons as a definitive host for this virus.
Gibbon ape leukemia virus and related viruses
Currently there are seven GALV or GALV-related strains with published proviral sequence. These are GALV-SEATO [7], GALV-SF [8], GALV-H [9], GALV-BR [10] GALV-mar (unpublished sequence GenBank: U20589.1), GALV-X [11] and Simian sarcoma-associated virus (SSAV) [12]. Most of the GALV strains were isolated from animals whose associated neoplastic syndromes were only recorded in specific cohorts from captive research institutes. GALV-mar and GALV-X were derived in vitro. Simian sarcoma-associated virus is a defective GALV recombinant isolated once only from a new world Woolly Monkey (Lagothrix spp.) that putatively had transient exposure to a GALV-infected captive gibbon. The discovery of a number of strains of GALV in gibbons in various locations raises the question of whether gibbons from the SEATO facility were exported to other facilities, and if so did this movement of animals unwittingly lead to the spread of GALV in animals that were harbouring subclinical infections. The only reference the authors can find relating to export of gibbons from the SEATO facility is from Wikileaks (https://wikileaks.org/plusd/cables/1974BANGKO17800_b.html). It refers to a shipment of 11 gibbons to the University of California, Davis in August 1973. It seems plausible that other shipments may have occurred as well and this could have allowed for the dissemination of the virus to other facilities.
The discovery of an apparently contagious oncogenic gammaretrovirus in higher order primates stimulated a great deal of research and one area of interest was the origin of this virus. Early reports suggested that GALV is closely related and possibly derived from Murine leukemia-like viruses (MuLV) hosted by South East Asian rodent species [13, 14]. The initial assessment of ‘close’ relationship between GALV and MuLV has progressively devolved toward the status of validated fact in some recent publications [15]. However, the initial ‘close’ relationship inference was derived from the results of low-resolution serological and DNA homology methods, including cross-reacting but poorly defined epitopes of p30. More recent work has questioned the validity of this inference based on viral receptor usage [16], and to our knowledge, almost all recent phylograms based on extended sequence alignments show the GALV-KoRV clade clearly polyphyletic with the combined MuLV/FeLV clades. Modern methods using direct sequence comparison against the putative “close relative” MuLV candidates for which sequences are available such as Mus dunni endogenous virus (MdEV) shows sequence identity of 68% for pol and 55% for env with the corresponding GALV-SEATO sequence, and for Mus caroli endogenous virus (McERV), the identities are 69% for pol and 55% for env.
Melomys burtoni retrovirus and its vertebrate host
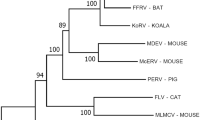
The recent discovery of proviral sequences in genomic DNA from a native Australian rodent, the grassland melomys (Melomys burtoni) that are remarkably similar to GALV, opens a new chapter in the quest for the origins of GALV [17], (Fig. 1). These partial proviral sequences, designated Melomys burtoni retrovirus (MbRV) have a high degree of identity with GALV. A high posterior probability for the ancestral placement of MbRV in our phylogeny supports the hypothesis that the MbRV–GALV common ancestor was basal to all known GALV viruses. However, we note that some caution is needed when interpreting these results, particularly with respect to the following caveats. First, low posterior probabilities for the placement of many GALV and WMV sequences indicate that the true relationships of these viruses are uncertain, perhaps due to different evolutionary histories and selection pressures acting on the pol and env genes. Second, although multi-species coalescent models outperform concatenation approaches to estimate relationships using multi-locus data, resolution of divergence times (and perhaps of topologies) will be limited without sampling multiple individuals per species.
Consensus multi-locus phylogeny showing evolutionary relationships of various gammaretroviruses. Analyses were performed using the *BEAST Bayesian algorithm, which co-estimates multiple gene trees within a shared species phylogenetic matrix. Untrimmed pol (3606 bp) and env (2391 bp) genes were included as separate alignments with unlinked substitution models (HKY+G for pol; GTR+I for env) and evolutionary rates (allowing the sampler to estimate strict clock rates from a Uniform (0 − 1*e100) prior distribution). The two gene partitions were embedded in a species tree matrix using a Yule speciation prior. Two MCMC chains were run for 40,000,000 iterations each, sampling every 2000 iterations (resulting in 30,000 posterior estimates after a 25% burnin). Chains were examined visually to confirm adequate mixing and ensure estimated parameter sample sizes were above 200. Colours of branches represent posterior probabilities of node placement, with warmer reds showing relatively low support and cooler blues showing high support. GenBank accession numbers for each taxa are as follows: KORVPciMCZ8574 (KF786282); KORVPcium3435 (KF786286); KORVPciBris (AF151794); KORVPcimaex1738 (KF786281); KORVPci582119 (KF786280); KORVKV522 (AB721500); KORVPciMCZ12454 (KF786283); KORVPciQMJ6480 (KF786284); KORVPciSN265 (KF786285); KORVBr2 (KC779547); WMVWD279 (KX059700); GALVSEATO (KT724048); WMVSSAV (KT724051); GALVH (KT724050); GALVSF (KT724047); GALVSFUMelb (X13194); GALVX (U60065); GALVBR (KT724049); MbRV (KF572483–KF572486); MDEV (AF053745); FELV (NC_001940)
Until the publication of these sequences, the virus most closely related to GALV was koala retrovirus (KoRV) [18]. KoRV, GALV and MbRV are all related, and it has been postulated that MbRV is the origin of KoRV given that koalas and the grassland melomys have overlapping geographical distributions [19, 20] and overlap in niche enough to reasonably postulate occasional cross-species viral transmission. What is intriguing however is that while MbRV and KoRV are clearly related averaging around 82% DNA identity across the polymerase loci, MbRV appears monophyletic with all the nominal strains of GALV (Fig. 1) showing up to 93% identity across aligned pol sequence. Currently, there are no published proviral sequences isolated from any species other than melomys which share this degree of identity with GALV.
Origin of GALV
The origin of GALV has been assumed to be a murine retrovirus derived from a local South East Asian rodent host. Melomys burtoni retrovirus would be a prime candidate for such an ancestor virus if its host, the grassland melomys, occurred in South East Asia and displayed some clear ecological overlap with gibbons. However, this species is restricted to north eastern Australia and Papua New Guinea and the genus Melomys is confined to Australasia east of Wallace’s line. Wallace’s line passes between the islands of Bali and Lombok in Indonesia. It divides the predominant fauna into that typical of South East Asia to the west and that typical of Australasia to the east. It is named after Alfred Wallace, a nineteenth century biologist who studied the fauna of the region [21]. No melomys species are known to occur west of Wallace’s line or in any part of mainland South East Asia [22]. Melomys most likely diverged from ancestral rodent stock and emerged in Australasia after the invasion and radiation of the Sahulian old endemic rodents around 5 million years ago [23]. It is likely then that any Melomys endogenous retroviruses (ERVs) would be proportionately divergent with the MuLV descendants found in extant mainland South East Asian rodents. To illustrate this point, the similarity between concatenated MbRV sequence (partial pol and env) obtained from a rodent east of Wallace’s line and corresponding sequence obtained from Mus dunni and Mus caroli (both species which occur on mainland South East Asia west of Wallace’s line) is in the order of 60%.
This opens the possibility that there is an as yet unknown retrovirus circulating in an Asian vertebrate host which shares a similar high degree of homology with GALV as does MbRV. The most likely species would appear to be either mainland South East Asian rodents or bats and two recent publications have put forward these alternative hypotheses. Brown and Tarlinton make the case for iatrogenic transmission of an ancestor GALV virus to gibbons from an as yet unidentified South East Asian rodent either at the SEATO facility or elsewhere in South East Asia [24]. They cite various AFRIMS reports supporting this claim. However, in a careful review of the cited reports, we can only find one reference where gibbons were inoculated with biologics of rodent origin and it is not clear that the rodent material was derived from a South East Asian rodent [25]. What makes the mainland South East Asian rodent theory problematic is that as noted above, almost all rodents have been geographically isolated to either South East Asia or Wallacia and Australasia, a division delineated by Wallace’s line, for millions of years. Given that exogenous retroviruses have a high mutation rate [26, 27] it seems unlikely that a putative ancestor virus to both MbRV and GALV could have maintained such a high degree of homology in separate vertebrate hosts which have been isolated for such a long period of time (Fig. 2).
Divergence of South East Asian and Australasian rodents: South East Asian rodents diverged from the Australasian old endemic rodents around 5 million years ago presumably after being isolated by sea level rise. Wallace’s line marks the geographic discontinuity that separates Asian from Australasian fauna. We propose that this also marked the co-divergence in murine ERV hosted by each group with Melomys eventually giving host to MbRV. Some Melomys are semi-arboreal raising the possibility of niche overlap and eventual cross-species transmission of MbRV to Koalas (Phascolarctos Spp.). This scenario implies that MbRV is a progenitor Murine-like ERV, which stabilised in the Melomys genome but retained infectivity over a few million years
Bats are another potential reservoir host and unlike rodents they can freely cross Wallace’s line and travel between South East Asia and Australasia [28]. However, the hypothesis that bats are the reservoir host also has some difficulties. For this to be true either there is a GALV strain circulating independently in bats and as we note above with respect to rodents it would be expected to have evolved with its host and no longer share such a high degree of homology with GALV. Alternatively, there may have been two host transmission events, from melomys to bats and then from bats to gibbons. We do not rule this hypothesis out but suggest it is an unlikely possibility.
Is the origin of GALV human after all?
An alternative theory explaining the origin of GALV presumes the possibility that MbRV or a closely related virus has been able to infect co-niched Sahul human hosts. In turn, then we speculate that biological material from these humans and hence contaminating MbRV virus was then iatrogenically inoculated into gibbons giving rise to GALV (Fig. 3). The rationale for this unlikely chain of events is based on the history of the early experimental work at the USA-SEATO medical research facility in Bangkok that used gibbons for Dengue Fever virus (DFV) and malaria research. Various strains of DFV were used, beginning in 1965/66, including Dengue-2 (N.G. “C”) (http://www.afrims.org/weblib/eapr/1966/APR66p015-041.pdf) and New Guin. “C” (Dengue -2) (http://www.afrims.org/weblib/eapr/1969/APR69p019-038.pdf).
Proposed transmission of MbRV from rodent to human to gibbon. In the 1960s, the US Army established a captive gibbon colony in Bangkok for experimental studies on human infectious diseases especially Dengue Fever and Malaria. In the early 1960s, Dengue Fever virus from infected people in Papua New Guinea was inoculated into captive gibbons at the Bangkok facility. This inoculation predated the first outbreak of GALV-associated lymphoid leukemia in gibbons at the colony. In addition, CNS tissue samples taken from residents with Kuru in the late 1960s were injected into gibbons at an experimental facility in the USA. Likewise, this inoculation predated the isolation of the GALV BR strain at that facility. We hypothesise that in each instance the human samples used to infect gibbons were contaminated with MbRV which in turn was the source of GALV. These scenarios imply that Papua New Guinea indigenous populations were either naturally infected with MbRV or that the blood samples taken from these people were cross contaminated with Melomys organic material. There are no records that either support or refute this direct iatrogenic mode of transmission for the remaining GALV strains (GALV SF, GALV X, GALV HI, SSAV and GALV Mar). Alternatively, the remaining strains could be secondary outbreaks independent of direct injection, but there are insufficient data to either rule in or rule out the wide variety of animal to animal transmission modes possible in the context of a disease with a long incubation period in high density experimental facilities
While the information in these reports is frustratingly incomplete, these references appear to refer to a New Guinea strain of DFV used to inoculate monkeys and gibbons. A later report mentions the “New Guinea “C” prototype virus” which is presumably the same virus mentioned in earlier studies (http://www.afrims.org/weblib/eapr/1969/APR69p196-197.pdf). Unfortunately, we have no more details on exactly how the DFV was obtained other than it originated from human patients. It is reasonable to assume that should a dengue fever patient have also been infected with a strain of MbRV that it is plausible that this virus could have also been transferred iatrogenically to gibbons. This report also notes that one of the gibbons inoculated with Dengue-2 strain subsequently developed leukemia. The first SEATO report which documents leukemia in gibbons is from 1968/69. Four gibbons are reported being diagnosed with leukemia over a 20-month period. Thus, the first apparent cases of leukemia at the SEATO facility occur some months or possibly years after the first inoculation of the New Guinea strain of DFV into gibbons (http://www.afrims.org/weblib/eapr/1969/APR69p196-197.pdf).
Another strain of GALV (GALV-BR) was isolated from gibbons at a research facility in the US. Initially five gibbons were imported from South East Asia in 1968. They were inoculated with brain material from Kuru patients from Papua New Guinea and died approximately 4 months later, in September 1968, from apparent pneumonia. Brain tissue from these gibbons was later co-cultivated with cell lines permissive to GALV, and GALV-BR was subsequently isolated from these cell cultures. The authors noted that “…contact of these gibbons with animals bearing known, experimentally induced infections with primate type C viruses was not possible” [10].
Thus, two distinct strains of GALV have been isolated from gibbons following the injection of material of human origin derived from Papua New Guinea. What makes this significant with respect to the putative origin of GALV is that the genus melomys, including M. burtoni, is widespread in Papua New Guinea [22]. Many people in Papua New Guinea live in remote or rural locations and have close contact with wildlife. In many areas, rodents are consumed as a source of protein.
We are aware that an MbRV-related virus (MelWMV) has recently been identified in a newly discovered sub species of melomys in the Indonesian Islands of Wallacia. While this intriguing new discovery adds to the narrative of MbRV and related viruses, this particular variant is not a candidate virus because it is an endogenous virus with premature stop codons and deletions [29]. In addition, it is not known to occur on mainland South East Asia [22].
It should be noted that in order for this hypothesis to be correct, it requires an infectious exogenous variant of MbRV to be circulating in rodents in Papua New Guinea and then to have passed to people via a cross-species transmission event. While we have partial proviral sequence from Australian melomys burtoni specimens, this does not guarantee the presence of infectious virus. We are currently pursuing further investigations to confirm the presence of an infectious virus in Papua New Guinea.
Is it possible that such a putative infectious strain of MbRV is circulating in Melomys species in Paua New Guinea and further that humans living in close contact with rodents in rural communities may have contracted this virus? Might this virus then have then been inadvertently passed on to gibbons when they were inoculated with biological material derived from human patients from Papua New Guinea? If such an iatrogenic infection occurred it would explain the sequence identity between GALV and MbRV.
If this hypothesis is proved correct and there is an infectious oncogenic human retrovirus circulating in Papua New Guinea, it has very significant health implications for at-risk populations. Such a virus may be present in humans as a subclinical infection and may not be as pathogenic or oncogenic as it is in gibbons. It is also possible that in remote regions of Papua New Guinea, people suffering from neoplastic disease caused by this putative virus might go undiagnosed and untreated.
The discovery of such a virus would open the door for novel anti-retroviral therapy for infected patients with neoplastic disease as well offering preventative measures such as possible vaccine development. It would also open the possibility that such a virus may be present in the human population in areas where Melomys burtoni occurs in Australia. People living in rural areas where the grassland melomys occurs could be exposed to this virus. Currently patients diagnosed with lymphoid tumours are not routinely screened for gammaretroviruses and such a putative virus could go undiagnosed in these patients. Extensive testing of human patients from various regions of Papua New Guinea, and in particular patients suffering from lymphoid tumours, as well as patients from the appropriate regions of Australia would shed further light on this hypothesis which has potentially important public health implications. We note that it is also possible although perhaps unlikely that the collection of samples from human patients may have been contaminated with infectious MbRV, thus allowing for the iatrogenic transmission to gibbons without human infection.
References
D.O. Johnsen, W.L. Wooding, P. Tanticharoenyos, C.H. Bourgeois, Malignant lymphoma in the gibbon. J. Am. Vet. Med. Assoc. 159, 563–566 (1971)
C.H. Lingeman, R.E. Reed, in Spontaneous hematopoietic neoplasms of non human primates. Review, case report and comparative studies, ed. by C.H. Lingeman and F.M. Garner, Symposium on comparative morphology of hematopoietic neoplasms (National Cancer Institute publishers, Washington D.C, 1969), pp. 157–167
A. De Paoli, F.M. Garner, Acute lymphocytic leukemia in a white-cheeked gibbon (Hylobates concolor). Cancer Res. 28, 2559–2561 (1968)
T.G. Kawakami et al., C-type virus associated with gibbon lymphosarcoma. Nat. New. Biol. 235, 170–171 (1972)
N. Teich, Taxonomy of retroviruses in RNA tumor viruses, Ed by R. Weiss, (Cold Spring Harbour Laborotory publishers, New York, 1984)
J.L. Siegal-Willott et al., Evaluation of captive gibbons (Hylobates spp., Nomascus spp., Symphalangus spp.) in North American Zoological Institutions for Gibbon Ape Leukemia Virus (GALV). J. Zoo Wildl. Med. 46, 27–33 (2015)
T.G. Kawakami, P.M. Buckley, Antigenic studies on gibbon type-C viruses. Transpl. Proc. 6, 193–196 (1974)
T.G. Kawakami, G.V. Kollias Jr., C. Holmberg, Oncogenicity of gibbon type-C myelogenous leukemia virus. Int. J. Cancer 25, 641–646 (1980)
J.M. Krakower, S.R. Tronick, R.E. Gallagher, R.C. Gallo, S.A. Aaronson, Antigenic characterization of a new gibbon ape leukemia virus isolate: seroepidemiologic assessment of an outbreak of gibbon leukemia. Int. J. Cancer 22, 715–720 (1978)
G.J. Todaro, M.M. Lieber, R.E. Benveniste, C.J. Sherr, Infectious primate type C viruses: three isolates belonging to a new subgroup from the brains of normal gibbons. Virology 67, 335–343 (1975)
G. Burtonboy, N. Delferriere, B. Mousset, M. Heusterspreute, Isolation of a C-type retrovirus from an HIV infected cell line. Arch. Virol. 130, 289–300 (1993)
G.H. Theilen, D. Gould, M. Fowler, D.L. Dungworth, C-type virus in tumor tissue of a woolly monkey (Lagothrix spp.) with fibrosarcoma. J. Natl. Cancer. Inst. 47, 881–889 (1971)
R. Callahan, C. Meade, G.J. Todaro, Isolation of an endogenous type C virus related to the infectious primate type C viruses from the Asian rodent Vandeleuria oleracea. J. Virol. 30, 124–131 (1979)
M.M. Lieber et al., Isolation from the asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc. Natl. Acad. Sci. USA 72, 2315–2319 (1975)
J. Denner, Endogenous retroviruses, in retroviruses, ed. by R. Kurth, N. Bannert (Caister Academic Press publishers, Norfolk, 2010)
A.D. Miller, U. Bergholz, M. Ziegler, C. Stocking, Identification of the myelin protein plasmolipin as the cell entry receptor for Mus caroli endogenous retrovirus. J. Virol. 82, 6862–6868 (2008)
G.S. Simmons, D. Clarke, J. McKee, P. Young, J. Meers, Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): a putative link between gibbon ape leukemia virus and koala retrovirus. PLOS ONE 9, e106954 (2014)
J.J. Hanger, L.D. Bromham, J.J. McKee, T.M. O’Brien, W.F. Robinson, The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J. Virol. 74, 4264–4272 (2000)
T.D. Redhead, Grassland melomys, in Complete book of australian mammals, ed. by R. Strahan (Angus and Roberston publishers, Melbourne, 1983), pp. 370–379
R.W. Martin, Koala, in Complete book of Australian mammals, ed. by R. Strahan (Angus and Robertson publishers, Melbourne, 1983)
J. Bastin, Introduction, in The Malay Archipeligo, ed. by A.R. Wallace (Oxford University Press Publishers, New York, 1989), pp. 7–27
Mosaic Tailed rats, in Walkers mammals of the world, vol. 2, ed. by M. Nowak, (Johns Hopkins University Press publishers, Baltimore, 1999), pp. 1548–1549
K.C. Rowe, M.L. Reno, D.M. Richmond, R.M. Adkins, S.J. Steppan, Pliocene colonization and adaptive radiations in Australia and New Guinea (Sahul): multilocus systematics of the old endemic rodents (Muroidea: Murinae). Mol. Phylogenet. Evol. 47, 84–101 (2008)
K. Brown, R.E. Tarlinton, Is gibbon ape leukaemia virus still a threat? Mamm. Rev. 47, 53–61 (2016)
P.C. Smith, L.C. Olson, T.M. Yuill, Experimental infections of gibbons with herpesvirus hominis SEATO medical research laboratory annual progress reports. US Army Medical Component (1969)
J. Overbaugh, C.R.M. Bangham, Selection forces and constraints on retroviral sequence variation. Science 292, 1106–1109 (2001)
L. Andréoletti, B. Réveil, H. Moret, V. Brodard, F. Philbert, T. Tabary, J.H.M. Cohen, Significant genetic and antigenic variability within the env gene of systemic human immunodeficiency virus type 1 group O populations during the natural course of a heterosexual infection: a pilot study. J. Clin. Micro. 45, 1319–1321 (2007)
J. Denner, Are bat retroviruses the origin of the gibbon ape leukaemia virus (GaLV) and the koala retrovirus (KoRV)? Viruses (2016). doi:10.3390/v8120336
N. Alfano et al., Endogenous Gibbon Ape Leukemia Virus Identified in a Rodent (Melomys burtoni subsp.) from Wallacia (Indonesia). J. Virol. 90, 8169–8180 (2016)
Acknowledgements
The authors wish to acknowledge the assistance of Gabriel Milinovich and Ricardo Soares Magalhaes in the preparation of this manuscript.
Author contributions
N. Clark constructed the phylogenetic tree in Fig. 1. F. Shapter assisted with the writing of the manuscript and interpretation of the phylogenetics.
Funding
This work was funded in part by the Australian Research Council Linkage Grant No. LP0453692. http://www.arc.gov.au/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest relating to this manuscript.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Additional information
Edited by Detlev H. Kruger.
J. McKee and G. Simmons contributed equally to the writing of this manuscript.
Rights and permissions
About this article
Cite this article
McKee, J., Clark, N., Shapter, F. et al. A new look at the origins of gibbon ape leukemia virus. Virus Genes 53, 165–172 (2017). https://doi.org/10.1007/s11262-017-1436-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1436-0