Abstract
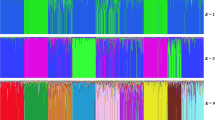
The Afrikaner is an indigenous South African breed of “Sanga” type beef cattle along with breeds such as the Drakensberger and Nguni. Six composite breeds have been developed from crosses with the Afrikaner. Additionally, Afrikaner has been the base from which exotic breeds were established in South Africa through backcrossing. The study examined genetic diversity of Afrikaner cattle by genotyping 1257 animals from 27 herds in different geographic areas of South Africa and Namibia using 11 microsatellite markers. Multiple-locus assignment, performed using the Bayesian clustering algorithm of STRUCTURE, revealed three underlying genotypic groups. These groups were not geographically localized. Across herds and markers, the proportion of unbiased heterozygosity ranged from 0.49 to 0.72 averaging 0.57; mean number of alleles per locus ranged from 3.18 to 7.09, averaging 4.81; and allelic richness ranged from 2.35 to 3.38, averaging 2.67. It is concluded that a low inbreeding level of 2.7% and a moderate to high degree of variation still persists within the Afrikaner cattle breed, despite the recent decline in numbers of animals.


Similar content being viewed by others
References
Bessa, I., Pinheiro, I., Matola, M., Dzama K., Rocha, A. and Alexandrino, P., 2009. Genetic diversity and relationships among indigenous Mozambican cattle breeds, South African Journal of Animal Science, 39, 61--72
Burrow, H.M., 1993. The Effects of Inbreeding in Beef Cattle. Animal Breeding Abstracts, 61, 11.
Carruthers, C.R., Plante, Y. and Schmutz, S.M., 2011. Comparison of Angus cattle populations using gene variants and microsatellites, Canadian Journal of Animal Science, 91, 81--85
Coetzer, W.A. and Van Marle, J., 1972. Die voorkoms van puberteit en daaropvolgende estrusperiode by vleisrasverse, South African Journal of Animal Science, 2, 17--18
Delgado, J.V., Martínez, A.M., Acosta, A., Álvarez, L.A., Armstrong, E., Camacho, E., Cañón, J., Cortés, O., Dunner, S., Landi, V. Marques, J.R., Martín-Burriel, I., Martínez, O.R., Martínez, R.D., Melucci, L., Muñoz, J.E., Penedo, M.C.T., Postiglioni, A., Quiróz, J., Rodellar, C., Sponenberg, P., Uffo, O., Ulloa-Arvizu, R., Vega-Pla, J.L., Villalobos, A., Zambrano, D., Zaragoza, P., Gama, L.T. and Ginja C., 2011. Genetic characterization of Latin-American Creole cattle using microsatellite markers, Animal Genetics, 43, 2--10
Earl, D.A. and vonHoldt, B.M., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method, Conservation Genetics Resources, 4, 359--361, ( http://taylor0.biology.ucla.edu/structureHarvester/ )
Evanno, G., Regnaut, S. and Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study, Molecular Ecology, 14, 2611--2620
Excoffier, L., Laval, G. and Schneider, S., 2005. Arlequin, version 3.0: an integrated software package for Population Genetics Data Analysis. Evolutionary bioinformatics online, 1, p.47.
Fan, B., Chen, Y.Z., Moran, C., Zhao, S.H., Liu, B., Yu, M., Zhu, M.J., Xiong, T.A. and Li, K., 2005. Individual-breed assignment analysis in swine populations by using microsatellite markers, Asian-Australasian Journal of Animal Sciences, 18, 1529--1534
FAO., 1998. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans. In: Management of Small Populations at Risk, Rome Italy, United Nations
Fernández, J., Villanueva, B., Pong-Wong, R. and Toro, M.A., 2005. Efficiency of the use of pedigree and molecular marker information in conservation programs, Genetics, 170, 1313--1321
Fernández, M.E., Goszczynski, D.E., Lirón, J.P., Villegas-Castagnasso, E.E., Carino, M.H., Ripoli, M.V., Rogberg-Muñoz, A., Posik, D.M., Peral-García, P. and Giovambattista, G., 2013. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd, Genetics and Molecular Biology, 36, 185--191
Frankham, R., Ballou, J.D. and Briscoe, D.A., 2002. Introduction to Conservation Genetics, (Cambridge University Press, UK)
Goudet, J., 2002. FSTAT, a program to estimate and test gene diversities and fixation indices version 2.9.3.2, http://www2.unil.ch/popgen/softwares/fstat.htm)
Hepsibha, P., Gogoi, A. and Karthickeyan, S.M.K., 2014. Molecular genetic architecture of Hallikar cattle (Bos indicus) revealed through microsatellite markers, Indian Journal of Biotechnology, 13, 411--413
Jordaan, F.J., 2015. Genetic and environmental trends in landrace beef breeds and the effect on cow productivity, (MSc dissertation, University of the Free State, South Africa)
Makina, S, O., Whitacre, L.K., Decker, J.E., Taylor, J.F., MacNeil, M.D., Scholtz, M.M., Muchadeyi, F.C., van Marle-Köster, E., Makgahlela, M.L. and Maiwashe A., 2016. Insight into the genetic composition of South African Sanga cattle using dense SNP data from cattle breeds worldwide, Genetics Selection Evolution, 48, 88
Ndiaye, N.D., Sow, A., Dayo, G-K., Ndiaye, S., Sawadogo G.J. and Sembène, M., 2015. Genetic diversity and phylogenetic relationships in local cattle breeds of Senegal based on autosomal microsatellite markers, Veterinary World, 8, 994--1005.
Nei, M., 1987. Molecular Evolutionary Genetics, (Columbia University Press, New York).
Newman, S., 2011. The role of genetic evaluation technology in enhancing global competitiveness. Beef Improvement Federation Research Symposium and Annual Meeting, Bozeman, Montana, 11--17
Park, S.D.E., 2001. Trypanotolerance in West African cattle and the population genetic effects of selection, (PhD thesis, Trinity College, University of Dublin)
Payne, W.J.A. and Wilson, T.R., 1999. An Introduction to Animal Husbandry in the Tropics, (Blackwell Science LTD, Oxford)
Pienaar, L., Neser, F.W.C., Grobler, J.P., Scholtz, M.M. and MacNeil, M.D., 2015. Pedigree analysis of the Afrikaner cattle breed, Animal Genetic Resources, 57, 51--56.
Pritchard, J.K., Stephens, M., Rosenberg, N.A. and Donnelly, P., 2000. Association mapping in structured populations, American Journal of Human Genetics, 67, 170--181
Rosenberg, N.A., Burke, T., Elo, K., Feldman, M.W., Freidlin, P.J., Groenen, M.A.M., Hillel, J., Mäki-Tanila, A., Tixier-Boichard, M., Vignal, A., Wimmers, K. and Weigend, S., 2001. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics, 159, 699--713
Schaid, D.J., Guenther, J.C., Christensen, G.B., Hebbring, S., Rosenow, C., Hilker, C.A., McDonnell, S.K., Cunningham, J.M., Slager, S.L., Blute, M.L. and Thibodeau, S.N., 2004. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer–susceptibility loci. American Journal of Human Genetics, 75, 948--965
Schlenker, W. and Lobell, D.B., 2010. Robust negative impacts of climate change on African agriculture, Environmental Research Letters, 5, 014010
Scholtz, M.M., 2005. The role of research and the seedstock industry in the in situ conservation of livestock genetic resources. The 4th All Africa Conference on Animal Agriculture, Arusha, Tanzania, 311--316.
Scholtz, M.M., 2010. Beef Breeding in South Africa, (Agricultural Research Council, Pretoria)
Scholtz, M.M., McManus, C., Leeuw, K.-J., Louvandini, H., Seixas, L., de Melo, C.B., Theunissen, A. and Neser, F.W.C., 2013. The effect of global warming on beef production in developing countries of the southern hemisphere. Natural Science, 5, 106--119
Steenkamp, J.J. and Tissier, L., 2016. The history of the Afrikaner Cattle Breeders’ Society of South Africa. In: M.M. Scholtz, F.W.C. Neser, and L. Tissier (eds), The Afrikaner breed of cattle: South Africa’s heritage for food security. ARC, Pretoria, 10--17
Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, A., 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0, Molecular Biology and Evolution, 30, 2725--2729
Weir, B.S. and Cockerham, C.C., 1984. Estimating F-Statistics for the analysis of population structure, Evolution, 38, 1358--1370
Wright, S., 1951. The genetical structure of populations, Annals of Eugenics, 15, 323--354
Zhang, G.X., Wang, Z.G., Chen, W.S., Wu, C.X., Han, X., Chang, H., Zan, L.S., Li, R.L., Wang, J.H., Song, W.T., Xu, G.F., Yang, H.J. and Luo, Y.F., 2007. Genetic diversity and population structure of indigenous yellow cattle breeds of China using 30 microsatellite markers, Animal Genetics, 38, 550—559
Acknowledgements
We acknowledge the staff at the Department of Genetics at the University of the Free State. Also to the Agricultural Research Council-Animal Production Institute (ARC-API) and Unistel Medical Laboratories (UML) for providing the DNA databases of the seedstock animals. Likewise to the Staff at the Animal Genetics Laboratory at the ARC-API for assistance during sample analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Pienaar, L., Grobler, J.P., Scholtz, M.M. et al. Genetic diversity of Afrikaner cattle in southern Africa. Trop Anim Health Prod 50, 399–404 (2018). https://doi.org/10.1007/s11250-017-1447-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-017-1447-9