Abstract
Background
An increasing number of systematic reviews (SRs) have evaluated the diagnostic values of next-generation sequencing (NGS) in infectious diseases (IDs).
Aim
This umbrella analysis aimed to assess the potential risk of bias in existing SRs and to summarize the published diagnostic values of NGS in different IDs.
Method
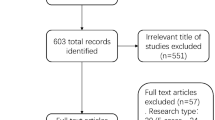
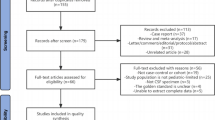
We searched PubMed, Embase, and the Cochrane Library until September 2023 for SRs assessing the diagnostic validity of NGS for IDs. Two investigators independently determined review eligibility, extracted data, and evaluated reporting quality, risk of bias, methodological quality, and evidence certainty in the included SRs.
Results
Eleven SRs were analyzed. Most SRs exhibited a moderate level of reporting quality, while a serious risk of bias was observed in all SRs. The diagnostic performance of NGS in detecting pneumocystis pneumonia and periprosthetic/prosthetic joint infection was notably robust, showing excellent sensitivity (pneumocystis pneumonia: 0.96, 95% CI 0.90–0.99, very low certainty; periprosthetic/prosthetic joint infection: 0.93, 95% CI 0.83–0.97, very low certainty) and specificity (pneumocystis pneumonia: 0.96, 95% CI 0.92–0.98, very low certainty; periprosthetic/prosthetic joint infection: 0.95, 95% CI 0.92–0.97, very low certainty). NGS exhibited high specificity for central nervous system infection, bacterial meningoencephalitis, and tuberculous meningitis. The sensitivity to these infectious diseases was moderate. NGS demonstrated moderate sensitivity and specificity for multiple infections and pulmonary infections.
Conclusion
This umbrella analysis indicates that NGS is a promising technique for diagnosing pneumocystis pneumonia and periprosthetic/prosthetic joint infection with excellent sensitivity and specificity. More high-quality original research and SRs are needed to verify the current findings.

Similar content being viewed by others
References
Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2095–128. https://doi.org/10.1016/S0140-6736(12)61728-0.
Wang X, Hong XZ, Li YW, et al. Microfluidics-based strategies for molecular diagnostics of infectious diseases. Mil Med Res. 2022;9(1):1–7. https://doi.org/10.1186/s40779-022-00374-3.
Ackerman CM, Myhrvold C, Thakku SG, et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–82. https://doi.org/10.1038/s41586-020-2279-8.
Zhang XX, Liu JS, Han LF, et al. Towards a global one health index: a potential assessment tool for one health performance. Infect Dis Poverty. 2022;11(1):57. https://doi.org/10.1186/s40249-022-00979-9.
Liu Q, Jing W, Liu M, et al. Health disparity and mortality trends of infectious diseases in BRICS from 1990 to 2019. J Glob Health. 2022;12:04028. https://doi.org/10.7189/jogh.12.04028.
Acharya S, Barber SL, Lopez-Acuna D, et al. BRICS and global health. Bull World Health Organ. 2014;92:386-386A. https://doi.org/10.2471/BLT.14.140889.
Summer A. IDS working paper: where do the world’s poor live? A new update. 2012. Available: https://www.ids.ac.uk/download.php?file=files/dm-file/Wp393.pdf. Accessed: 20 Sept 2021.
GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204–22. https://doi.org/10.1016/S0140-6736(20)30925-9.
Huang G, Guo F. Loss of life expectancy due to respiratory infectious diseases: findings from the global burden of disease study in 195 countries and territories 1990–2017. J Popul Res (Canberra). 2022;39(1):1–43. https://doi.org/10.1007/s12546-021-09271-3.
Springer BD, Cahue S, Etkin CD, et al. Infection burden in total hip and knee arthroplasties: an international registry-based perspective. Arthroplast Today. 2017;3(2):137–40. https://doi.org/10.1016/j.artd.2017.05.003.
Rietbergen L, Kuiper JW, Walgrave S, et al. Quality of life after staged revision for infected total hip arthroplasty: a systematic review. Hip Int. 2016;26(4):311–8. https://doi.org/10.5301/hipint.5000416.
Bozic KJ, Kurtz SM, Lau E, et al. The epidemiology of revision total knee arthroplasty in the United States. Clin Orthop Relat Res. 2010;468(1):45–51. https://doi.org/10.1007/s11999-009-0945-0.
Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–96. https://doi.org/10.1097/01.CCM.0000217961.75225.E9.
Glimåker M, Johansson B, Grindborg Ö, et al. Adult bacterial meningitis: earlier treatment and improved outcome following guideline revision promoting prompt lumbar puncture. Clin Infect Dis. 2015;60:1162–9. https://doi.org/10.1093/cid/civ011.
Weiss SL, Fitzgerald JC, Balamuth F, et al. Delayed antimicrobial therapy increases mortality and organ dysfunction duration in pediatric sepsis. Crit Care Med. 2014;42:2409–17. https://doi.org/10.1097/CCM.0000000000000509.
Erickson TA, Muscal E, Munoz FM, et al. Infectious and autoimmune causes of encephalitis in children. Pediatrics. 2020;145(6): e20192543. https://doi.org/10.1542/peds.2019-2543.
Messacar K, Parker SK, Todd JK, et al. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol. 2017;55:715–23. https://doi.org/10.1128/JCM.02264-16.
Haidar G, Singh N. Fever of unknown origin. N Engl J Med. 2022;386(5):463–77. https://doi.org/10.1056/NEJMra2111003.
Heesterbeek H, Anderson RM, Andreasen V, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347(6227):aaa4339. https://doi.org/10.1126/science.aaa4339.
Denny KJ, De Waele J, Laupland KB, et al. When not to start antibiotics: avoiding antibiotic overuse in the intensive care unit. Clin Microbiol Infect. 2020;26(1):35–40. https://doi.org/10.1016/j.cmi.2019.07.007.
Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67:S231–40. https://doi.org/10.1093/cid/ciy693.
Peng JM, Du B, Qin HY, et al. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J Infect. 2021;82:22–7. https://doi.org/10.1016/j.jinf.2021.01.029.
Yoon HK, Cho SH, Lee DY, et al. A review of the literature on culture-negative periprosthetic joint infection: epidemiology, diagnosis and treatment. Knee Surg Relat Res. 2017;29(3):155–64. https://doi.org/10.5792/ksrr.16.034.
Ji XC, Zhou LF, Li CY, et al. Reduction of human DNA contamination in clinical cerebrospinal fluid specimens improvesthe sensitivity of metagenomic next-generation sequencing. J Mol Neurosci. 2020;70(5):659–66. https://doi.org/10.1007/s12031-019-01472-z.
Fei X, Li C, Zhang Y, et al. Next-generation sequencing of cerebrospinal fluid for the diagnosis of neurocysticercosis. Clin Neurol Neurosurg. 2020;193: 105752. https://doi.org/10.1016/j.clineuro.2020.105752.
Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14:319–38. https://doi.org/10.1146/annurev-pathmechdis-012418-012751.
Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–17. https://doi.org/10.1056/NEJMoa1401268.
Yu X, Jiang W, Shi Y, et al. Applications of sequencing technology in clinical microbial infection. J Cell Mol Med. 2019;23(11):7143–50. https://doi.org/10.1111/jcmm.14624.
To RK, Ramchandar N, Gupta A, et al. Use of plasma metagenomic next-generation sequencing for pathogen identification in pediatric endocarditis. Pediatr Infect Dis J. 2021;40(5):486–8. https://doi.org/10.1097/INF.0000000000003038.
Lieberman JA, Naureckas Li C, Lamb GS, et al. Case report: comparison of plasma metagenomics to bacterial PCR in a case of prosthetic valve endocarditis. Front Pediatr. 2020;8:575674. https://doi.org/10.3389/fped.2020.575674.
Farnaes L, Wilke J, Ryan Loker K, et al. Community-acquired pneumonia in children: cell-free plasma sequencing for diagnosis and management. Diagn Microbiol Infect Dis. 2019;94:188–91. https://doi.org/10.1016/j.diagmicrobio.2018.12.016.
Wang J, Han Y, Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 2019;19(1):252. https://doi.org/10.1186/s12890-019-1022-4.
Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–40. https://doi.org/10.1056/NEJMoa1803396.
Cai Y, Fang X, Chen Y, et al. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int J Infect Dis. 2020;96:573–8. https://doi.org/10.1016/j.ijid.2020.05.125.
Fang X, Cai Y, Shi T, et al. Detecting the presence of bacteria in low-volume preoperative aspirated synovial fluid by metagenomic next-generation sequencing. Int J Infect Dis. 2020;99:108–16. https://doi.org/10.1016/j.ijid.2020.07.039.
Kildow BJ, Ryan SP, Danilkowicz R, et al. Next-generation sequencing not superior to culture in periprosthetic joint infection diagnosis. Bone Joint J. 2021;103:26–31. https://doi.org/10.1302/0301-620X.103B1.BJJ-2020-0017.R3.
Tarabichi M, Shohat N, Goswami K, et al. Diagnosis of periprosthetic joint infection: the potential of next-generation sequencing. J Bone Joint Surg Am. 2018;100:147–54. https://doi.org/10.2106/JBJS.17.00434.
Tarabichi M, Alvand A, Shohat N, et al. Diagnosis of Streptococcus canis periprosthetic joint infection: the utility of next-generation sequencing. Arthroplast Today. 2018;4:20–3. https://doi.org/10.1016/j.artd.2017.08.005.
Tarabichi M, Shohat N, Goswami K, et al. Can next generation sequencing play a role in detecting pathogens in synovial fluid? Bone Joint J. 2018;100:127–33. https://doi.org/10.1302/0301-620X.100B2.BJJ-2017-0531.R2.
Torchia MT, Amakiri I, Werth P, et al. Characterization of native knee microorganisms using next-generation sequencing in patients undergoing primary total knee arthroplasty. Knee. 2020;27:1113–9. https://doi.org/10.1016/j.knee.2019.12.013.
Goggin KP, Gonzalez-Pena V, Inaba Y, et al. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol. 2020;6(4):552–6. https://doi.org/10.1001/jamaoncol.2019.4120.
Eichenberger EM, de Vries CR, Ruffin F, et al. Microbial cell-free DNA identifies etiology of bloodstream infections, persists longer than conventional blood cultures, and its duration of detection is associated with metastatic infection in patients with staphylococcus aureus and gram-negative bacteremia. Clinic Infect Dis. 2022;74(11):2020–7. https://doi.org/10.1093/cid/ciab742.
Gosiewski T, Ludwig-Galezowska AH, Huminska K, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method-the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36(2):329–36. https://doi.org/10.1007/s10096-016-2805-7.
Grumaz S, Grumaz C, Vainshtein Y, et al. Enhanced performance of next-generation sequencing diagnostics compared with standard of care microbiological diagnostics in patients suffering from septic shock. Crit Care Med. 2019;47:e394-402. https://doi.org/10.1097/CCM.0000000000003658.
Zhang HC, Ai JW, Cui P, et al. Incremental value of metagenomic next generation sequencing for the diagnosis of suspected focal infection in adults. J Infect. 2019;79:419–25.
Hong DK, Blauwkamp TA, Kertesz M, et al. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn Microbiol Infect Dis. 2018;92(3):210–3. https://doi.org/10.1016/j.diagmicrobio.2018.06.009.
Armstrong AE, Rossoff J, Hollemon D, et al. Cell-free DNA next-generation sequencing successfully detects infectious pathogens in pediatric oncology and hematopoietic stem cell transplant patients at risk for invasive fungal disease. Pediatr Blood Cancer. 2019;66: e27734. https://doi.org/10.1002/pbc.27734.
Yan L, Sun W, Lu Z, et al. Metagenomic next-generation Sequencing (mNGS) in cerebrospinal fluid for rapid diagnosis of tuberculosis meningitis in HIV-negative population. Int J Infect Dis. 2020;96:270–5. https://doi.org/10.1016/j.ijid.2020.04.048.
Fung M, Zompi S, Seng H, et al. Plasma cell-free DNA next-generation sequencing to diagnose and monitor infections in allogeneic hematopoietic stem cell transplant patients. Open Forum Infect Dis. 2018;5:ofy301. https://doi.org/10.1093/ofid/ofy301.
Zhou Y, Hemmige V, Dalai SC, et al. Utility of whole-genome next-generation sequencing of plasma in identifying opportunistic infections in HIV/AIDS. Open AIDS J. 2019;13:7–11.
Chen S, Kang Y, Li D, et al. Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in patients with pulmonary infections: systematic review and meta-analysis. Int J Infect Dis. 2022;122:867–73. https://doi.org/10.1016/j.ijid.2022.07.054.
Tekin A, Truong HH, Rovati L, et al. The diagnostic accuracy of metagenomic next-generation sequencing in diagnosing pneumocystis pneumonia: a systemic review and meta-analysis. Open Forum Infect Dis. 2023;10(9):442. https://doi.org/10.1093/ofid/ofad442.
Qu C, Chen Y, Ouyang Y, et al. Metagenomics next-generation sequencing for the diagnosis of central nervous system infection: a systematic review and meta-analysis. Front Neurol. 2022;13: 989280. https://doi.org/10.3389/fneur.2022.989280.
Kanaujia R, Biswal M, Angrup A, et al. Diagnostic accuracy of the metagenomic next-generation sequencing (mNGS) for detection of bacterial meningoencephalitis: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2022;41(6):881–91. https://doi.org/10.1007/s10096-022-04445-0.
Yu G, Zhao W, Shen Y, et al. Metagenomic next generation sequencing for the diagnosis of tuberculosis meningitis: a systematic review and meta-analysis. PLoS ONE. 2020;15(12): e0243161. https://doi.org/10.1371/journal.pone.0243161.
Li M, Zeng Y, Wu Y, et al. Performance of sequencing assays in diagnosis of prosthetic joint infection: a systematic review and meta-analysis. J Arthroplasty. 2019;34(7):1514-1522.e4. https://doi.org/10.1016/j.arth.2019.02.044.
Tan J, Liu Y, Ehnert S, et al. The effectiveness of metagenomic next-generation sequencing in the diagnosis of prosthetic joint infection: a systematic review and meta-analysis. Front Cell Infect Microbiol. 2022;12: 875822. https://doi.org/10.3389/fcimb.2022.875822.
Tang Y, Zhao D, Wang S, et al. Diagnostic value of next-generation sequencing in periprosthetic joint infection: a systematic review. Orthop Surg. 2022;14(2):190–8. https://doi.org/10.1111/os.13191.
Cheok T, Smith T, Siddiquee S, et al. Synovial fluid calprotectin performs better than synovial fluid polymerase chain reaction and interleukin-6 in the diagnosis of periprosthetic joint infection : a systematic review and meta-analysis. Bone Joint J. 2022;104(3):311–20. https://doi.org/10.1302/0301-620X.104B3.BJJ-2021-1320.R1.
Hantouly AT, Alzobi O, Toubasi AA, et al. Higher sensitivity and accuracy of synovial next-generation sequencing in comparison to culture in diagnosing periprosthetic joint infection: a systematic review and meta-analysis. Knee Surg Sports Traumatol Arthrosc. 2023;31(9):3672–83. https://doi.org/10.1007/s00167-022-07196-9.
Aromataris E, Fernandez R, Godfrey CM, et al. Summarizing systematic reviews: methodological development, conduct and reporting of an umbrella review approach. Int J Evid Based Healthc. 2015;13(3):132–40. https://doi.org/10.1097/XEB.0000000000000055.
Gates M, Gates A, Pieper D, et al. Reporting guideline for overviews of reviews of healthcare interventions: development of the PRIOR statement. BMJ. 2022;9(378): e070849. https://doi.org/10.1136/bmj-2022-070849.
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;21(339): b2700. https://doi.org/10.1136/bmj.b2700.
Whiting P, Savović J, Higgins JP, et al. A new tool to assess risk of bias in systematic reviews was developed. J Clin Epidemiol. 2016;69:225–34. https://doi.org/10.1016/j.jclinepi.2015.06.005.
Huan T, Yang L, An P, et al. Interpretation of quality evaluation tool AMSTAR 2 for systematic evaluation of randomized and nonrandomized preventive studies. Chin J Evid-Based Med. 2018;18(01):101–8. https://doi.org/10.7507/1672-2531.201711005.
Zhang F, Shen A, Zeng X, et al. Interpretation of systematic evaluation methodology quality evaluation tool AMSTAR 2. Chin J Evid-Based Cardiovasc Med. 2018;10(01):14–8. https://doi.org/10.3969/j.issn.1674-4055.2018.01.03.
Shea BJ, Reeves BC, Wells G, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. 2017;358: j4008. https://doi.org/10.1136/bmj.j4008.
Hultcrantz M, Mustafa RA, Leeflang MMG, et al. Defining ranges for certainty ratings of diagnostic accuracy: a GRADE concept paper. J Clin Epidemiol. 2020;117:138–48. https://doi.org/10.1016/j.jclinepi.2019.05.002.
Guyatt G, Oxman AD, Akl EA, et al. GRADE guidelines: 1. Introduction-GRADE evidence profiles and summary of findings tables. J Clin Epidemiol. 2011;64(4):383–94. https://doi.org/10.1016/j.jclinepi.2010.04.026.
Honghao L, Qiuyu Y, Mingyao S, et al. Advance in the GRADE approach to grade evidence from a systematic review of single diagnostic test accuracy. Chin J Evid Based Med. 2022;22(9):1090–8. https://doi.org/10.1007/s00259-022-05769-x.
Atkins D, Best D, Briss PA, et al. Grading quality of evidence and strength of recommendations. BMJ. 2004;328(7454):1490. https://doi.org/10.1136/bmj.328.7454.1490.
Liu J, Zhang Q, Dong YQ, et al. Diagnostic accuracy of metagenomic next-generation sequencing in diagnosing infectious diseases: a meta-analysis. Sci Rep. 2022;12(1):21032. https://doi.org/10.1038/s41598-022-25314-y.
Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–36. https://doi.org/10.7326/0003-4819-155-8-201110180-00009.
Lv M, Zhu C, Zhu C, et al. Clinical values of metagenomic next-generation sequencing in patients with severe pneumonia: a systematic review and meta-analysis. Front Cell Infect Microbiol. 2023;13:1106859. https://doi.org/10.3389/fcimb.2023.1106859.
The Editor Board of Chinese Journal of Infectious Diseases. Expert consensus on the clinical application of metagenomics next-generation sequencing on pathogen detection of infectious diseases. Chin J Infect Dis. 2020;38(11):681–9.
Clinical Microbiology Group of Chinese Society of Laboratory Medicine, Clinical Microbiology Group of Chinese Society of Microbiology and Immunology, and Society of Clinical Microbiology and Infection of China International Exchange and Promotion Association for Medical and Healthcare. Expert consensus on metagenomics next-generation sequencing application on pathogen detection of infectious diseases. Chin J Lab Med. 2021; 44(2):107–20.
The Society of Pediatrics of Chinese Medical Association, the Editor Board of Chinese Journal of Pediatrics. The Expert consensus on clinical of invasive pulmonary fungal infections in children. Chin J Pediatr. 2022;60(4):274–82. https://doi.org/10.3760/cma.j.cn112140-20220210-00112.
Infectious Disease Group of Respiratory Disease Branch of Chinese Medical Association. Guidelines for the diagnostic and treatment of adults with hospital-acquired pneumonia and ventilator-associated pneumonia in China. Chin J Tuberc Respir Dis. 2018;41(4):255–78.
The Subspecialty Group of Neurology of the Society of Pediatrics of Chinese Medical Association. Expert consensus on diagnosis and treatment of community acquired bacterial meningitis in children. Chin J Pediatr. 2019;57(8):584–291. https://doi.org/10.3760/cma.j.issn.0578-1310.2019.08.003.
Brown JR, Bharucha T, Breuer J. Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect. 2018;76(3):225–40. https://doi.org/10.1016/j.jinf.2017.12.014.
Kato H, Hagihara M, Asai N, et al. Comparison of microbial detection rates in microbial culture methods versus next-generation sequencing in patients with prosthetic joint infection: a systematic review and meta-analysis. J Orthop Surg Res. 2023;18(1):604. https://doi.org/10.1186/s13018-023-03973-5.
Lee RA, Al Dhaheri F, Pollock NR, et al. Assessment of the clinical utility of plasma metagenomic next-generation sequencing in a pediatric hospital population. J Clin Microbiol. 2020;58(7):e00419-420. https://doi.org/10.1128/JCM.00419-20.
Rossoff J, Chaudhury S, Soneji M, et al. Noninvasive diagnosis of infection using plasma next-generation sequencing: a single-center experience. Open Forum Infect Dis. 2019;6:327. https://doi.org/10.1093/ofid/ofz327.
Hogan CA, Yang S, Garner OB, et al. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis. 2021;72(2):239–45. https://doi.org/10.1093/cid/ciaa035.
Xu C, Chen X, Zhu G, et al. Utility of plasma cell-free DNA next-generation sequencing for diagnosis of infectious diseases in patients with hematological disorders. J Infect. 2023;86(1):14–23. https://doi.org/10.1016/j.jinf.2022.11.020.
Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–40. https://doi.org/10.1056/NEJMoa1803396.
NCT05290454: https://clinicaltrials.gov/study/NCT05290454. Accessed 15 Nov 2023.
NCT05979350: https://clinicaltrials.gov/study/NCT05979350. Accessed 15 Nov 2023.
NCT03884881: https://clinicaltrials.gov/study/NCT03884881. Accessed 15 Nov 2023.
NCT04946682 https://clinicaltrials.gov/study/NCT04946682. Accessed 15 Nov 2023.
Brenner T, Skarabis A, Stevens P, et al. Optimization of sepsis therapy based on patient-specific digital precision diagnostics using next generation sequencing (DigiSep-Trial)-study protocol for a randomized, controlled, interventional, open-label, multicenter trial. Trials. 2021;22(1):714. https://doi.org/10.1186/s13063-021-05667-x.
NCT05887037 https://clinicaltrials.gov/study/NCT05887037. Accessed 15 Nov 2023.
Perry R, Whitmarsh A, Leach V, et al. A comparison of two assessment tools used in overviews of systematic reviews: ROBIS versus AMSTAR-2. Syst Rev. 2021;10(1):273. https://doi.org/10.1186/s13643-021-01819-x.
Pieper D, Puljak L, González-Lorenzo M, et al. Minor differences were found between AMSTAR 2 and ROBIS in the assessment of systematic reviews including both randomized and nonrandomized studies. J Clin Epidemiol. 2019;108:26–33. https://doi.org/10.1016/j.jclinepi.2018.12.004.
Agudelo-Pérez S, Fernández-Sarmiento J, Rivera León D, et al. Metagenomics by next-generation sequencing (mNGS) in the etiological characterization of neonatal and pediatric sepsis: a systematic review. Front Pediatr. 2023;30(11):1011723. https://doi.org/10.3389/fped.2023.1011723.
Szlachta-McGinn A, Douglass KM, Chung UYR, et al. Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: a systematic review and meta-analysis. Eur Urol Open Sci. 2022;44:113–24. https://doi.org/10.1016/j.euros.2022.08.009.
Haddad SF, DeSimone DC, Chesdachai S, et al. Utility of metagenomic next-generation sequencing in infective endocarditis: a systematic review. Antibiotics (Basel). 2022;11(12):1798. https://doi.org/10.3390/antibiotics11121798.
Acknowledgements
We thank Juniie Lan, Jiaxue Wang, Shuimei Sun, Songsong Tan, and Jinyong Cao at Guizhou Provincia People's Hospital for helping with English language and manuscript editing.
Funding
This research received support from the National Natural Science Foundation of China (No. 72064004), the Hospital Pharmaceutical Research Project funded by the China Pharmaceutical Association (CPA-Z05-ZC-2023-002), and the Health Commission Foundation of Guizhou Province (gzwkj2022-224).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, H., Chen, Y., Ge, L. et al. An umbrella review of the diagnostic value of next-generation sequencing in infectious diseases. Int J Clin Pharm (2024). https://doi.org/10.1007/s11096-024-01704-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11096-024-01704-2