Summary
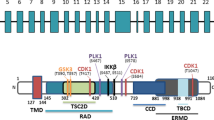
Tuberous sclerosis, neurological genetic disorder characterized by the formation of benign tumors or hamartomas in multiple organ systems, is recently getting much attention. Numerous papers describe still-not-fully-explained pathogenesis of the disease. Studies on tuberous sclerosis allowed identification of two tumor suppressor genes, TSC1 and TSC2, encoding proteins implicated in the disease: hamartin and tuberin, respectively. The importance of these proteins is confirmed by their ubiquitous character and by the fact that TSC1/TSC2 complex is involved in the regulation of the activity of mTOR, a master controller of protein translation. Thus, the meaning of hamartin and tuberin goes far beyond tuberous sclerosis. As far as the influence of the TSC1/TSC2 complex on protein translation is well described in numerous reviews, little attention is drawn to the recently discovered role of the TSC1/TSC2 complex in gene transcription via the WNT signaling pathway. The present paper focuses on recent developments documenting the role of hamartin and tuberin in the WNT pathway.
Similar content being viewed by others
References
Soucek T, Pusch O, Wienecke R, DeClue JE, Hengstschlager M: Role of the tuberous sclerosis gene-2 product in cell cycle control. Loss of the tuberous sclerosis gene-2 induces quiescent cells to enter S phase J Biol Chem 272: 29301–29308, 1997
Marino S: Medulloblastoma: developmental mechanisms out of control Trends Mol Med 11: 17–22, 2005
Li F, Chong ZZ, Maiese K: Vital elements of the Wnt-Frizzled signaling pathway in the nervous system Curr Neurovasc Res 2: 331–340, 2005
Bhanot P, Brink M, Samos CH, Hseih JC, Wang W, Macke JP, Andew D, Nathans J, Nusse R: A new member of the frizzled family from Drosophila functions as a wingless receptor Nature 282: 225–230, 1996
Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT: A frizzled homolog functions in a vertebrate Wnt signaling pathway Curr Biol 6: 1302–1306, 1996
Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E: Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity Dev Cell 4: 407–418, 2003
Thomas GM, Frame S, Goedert M, Nathke I, Polakis P, Cohen P: A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of axin, beta-catenin FEBS Lett 458:247–251, 1999
Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W: Functional interaction of beta-catenin with the transcription factor LEF-1 Nature 382:638–642, 1996
Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H: XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos Cell 86:391–399, 1996
Sakanaka C: Phosphorylation, regulation of beta-catenin by casein kinase I epsilon J Biochem (Tokyo) 132:697–703, 2002
Willert K, Brink M, Wodarz A, Varmus H, Nusse R: Casein kinase 2 associates with, phosphorylates dishevelled EMBO J 16:3089–3096, 1997
Bek S, Kemler R: Protein kinase CKII regulates the interaction of beta-catenin with alpha-catenin and its protein stability J Cell Sci 115: 4743–4753, 2002
Kowalczyk AP, Reynolds AB: Protecting your tail: regulation of cadherin degradation by p120-catenin Curr Opin Cell Biol 16: 522–527, 2004
Lilien J, Balsamo J, Arregui C, Xu G: Turn-off, drop-out: functional state switching of cadherins Dev Dyn 224: 18–29, 2002
Lilien J, Balsamo J: The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin Curr Opin Cell Biol 17: 459–465, 2005
Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K: The NF1 tumor suppressor critically regulates TSC2 and mTOR Proc Natl Acad Sci USA 102: 8573–8578, 2005
Koul D, Shen R, Bergh S, Lu Y, de Groot JF, Liu TJ, Mills GB, Yung WK: Targeting integrin-linked kinase inhibits Akt signaling pathways and decreases tumor progression of human glioblastoma Mol Cancer Ther 4: 1681–1688, 2005
Aldosari N, Bigner SH, Burger PC, Becker L, Kepner JL, Friedman HS, McLendon RE: MYCC and MYCN oncogene amplification in medulloblastoma. A fluorescence in situ hybridization study on paraffin sections from the Children’s Oncology Group Arch Pathol Lab Med 126: 540–544, 2002
Leonard JR, Cai DX, Rivet DJ, Kaufman BA, Park TS, Levy BK, Perry A: Large cell/anaplastic medulloblastomas and medullomyoblastomas: clinicopathological and genetic features J Neurosurg 95: 82–88, 2001
Reardon DA, Jenkins JJ, Sublett JE, Burger PC, Kun LK: Multiple genomic alterations including N-myc amplification in a primary large cell medulloblastoma Pediatr Neurosurg 32: 187–191, 2000
Jozwiak J: Hamartin and tuberin: working together for tumor formation Int J Canc 118: 1–5, 2006
Grolleau A, Bowman J, Pradet-Balade B, Puravs E, Hanash S, Garcia-Sanz JA, Beretta L: Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics J Biol Chem 277: 22175–22184, 2002
Knudson AG: Mutation and cancer: statistical study of retinoblastoma Proc Natl Acad Sci USA 68: 820–823, 1971
Henske EP, Wessner LL, Golden J, Scheithauer BW, Vortmeyer AO, Zhuang Z, Klein-Szanto AJ, Kwiatkowski DJ, Yeung RS: Loss of tuberin in both subependymal giant cell astrocytomas and angiomyolipomas supports a two-hit model for the pathogenesis of tuberous sclerosis tumors Am J Pathol 151: 1639–1647, 1997
Jozwiak S, Dabora S, Kasprzyk-Obara J, Domanska-Pakiela D, Grajkowska W: Tests for loss of heterozygosity in tuberous sclerosis Przegl Lek 58 Suppl. 1: 12–15, 2001
Knowles MA, Habuchi T, Kennedy W, Cuthbert-Heavens D: Mutation spectrum of the 9q34 tuberous sclerosis gene TSC1 in transitional cell carcinoma of the bladder Cancer Res 63: 7652–7656, 2003
Meikle L, McMullen JR, Sherwood MC, Lader AS, Walker V, Chan JA, Kwiatkowski DJ: A mouse model of cardiac rhabdomyoma generated by loss of Tsc1 in ventricular myocytes Hum Mol Genet 14: 429–435, 2005
Niida Y, Stemmer-Rachamimov AO, Logrip M, Tapon D, Perez R, Kwiatkowski DJ, Sims K, MacCollin M, Louis DN, Ramesh V: Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions Am J Hum Genet 69: 493–503, 2001
Ramesh V: Aspects of tuberous sclerosis complex (TSC) protein function in the brain Biochem Soc Trans 31: 579–583, 2003
Jozwiak J, Jozwiak S: Giant cells: contradiction to two-hit model of tuber formation? Cell Mol Neurobiol 25: 793–803, 2005
Kenerson HL, Aicher LD, True LD, Yeung RS: Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors Cancer Res 62: 5645–5650, 2002
Tetsu O, McCormick F: Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells Nature 398: 422–426, 1999
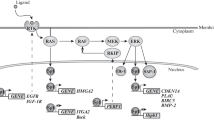
Mak BC, Takemaru K, Kenerson HL, Moon RT, Yeung RS: The tuberin-hamartin complex negatively regulates beta-catenin signaling activity J Biol Chem 278: 5947–5951, 2003
Mak BC, Kenerson HL, Aicher LD, Barnes EA, Yeung RS: Aberrant beta-catenin signaling in tuberous sclerosis Am J Pathol 167: 107–116, 2005
Rosner M, Freilinger A, Lubec G, Hengstschlager M: The tuberous sclerosis genes, TSC1 and TSC2, trigger different gene expression responses Int J Oncol 27, 1411–1424, 2005
Dabora SL, Jozwiak S, Franz DN, Roberts PS, Nieto A, Chung J, Choy YS, Reeve MP, Thiele E, Egelhoff JC, Kasprzyk-Obara J, Domanska-Pakiela D, Kwiatkowski DJ: Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs Am J Hum Genet 68: 64–80, 2001
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jozwiak, J., Wlodarski, P. Hamartin and tuberin modulate gene transcription via β-catenin. J Neurooncol 79, 229–234 (2006). https://doi.org/10.1007/s11060-006-9134-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-006-9134-0