Abstract
Authentication of seed provenance is an importance issue to avoid the negative impact of poor adaptation of progenies when planted outside their natural environmental conditions. The objective of this study was to evaluate the potential of near infrared (NIR) spectroscopy as rapid and non-destructive method for authentication of Picea abies L. Karst seed provenances. For this purpose, five seed lots from Sweden, Finland, Poland and Lithuania each were used. NIR reflectance spectra were recorded on individual seeds (n = 150 seeds × 5 seed lots × 4 provenances = 3000 seeds) using XDS Rapid Content Analyzer from 780 to 2500 nm with a resolution of 0.5 nm. Classification model was developed by orthogonal projection to latent structures-discriminant analysis. The performance of the computed classification model was validated using two test sets—internal (the same seed lots as the model but excluded during model development; n = 600 seeds) and external (seed lots not included in the model; n = 1158 seeds). For the internal test, the model correctly recognized 99% of Swedish, Finnish and Polish samples and 97% of Lithuanian seeds. For the external test samples, the model correctly assigned 81% of Swedish, 96% of Finnish, 98% of Lithuanian and 93% of Polish seeds to their respective classes. The mean classification accuracy was 99 and 95% for internal and external test set, respectively. The spectral differences among seed lots were attributed to differences in chemical composition of seeds, presumably fatty acids and proteins, which are the dominant storage reserves in P. abies seeds. In conclusion, the results demonstrate that NIR spectroscopy is a very promising method for monitoring putative seed provenances and in seed certification.
Similar content being viewed by others
Introduction
Picea abies L. Karst (Norway spruce) is widely distributed in northern and central Europe where its stands are managed mainly for timber production. It accounts for 42% of the total forest cover of Sweden (Statistical Yearbook of Forestry 2011), 23.1% of Lithuania (Vitas 2004), more than 33% of Finland (Ge et al. 2011) and 47% of Norway (Norwegian Ministry of the Environment 2009). It is considered to be the second ranking tree species of the Polish forests (Koprowski and Zielski 2006) with about 7% coverage. Among other things, sustainable forestry requires a continuous supply of high quality regeneration material, and seeds are the most commonly used regeneration material for reforestation. According to the Swedish Forest Agency, the total number of tree seedlings planted in Sweden in the year 2011 was 384 million, of which Norway spruce accounted for 225 million. However, annual seed production by P. abies trees is generally erratic between years due mainly to the paucity and periodicity of flowering (good flowering years followed by several poor years with little or no flowering). In Sweden, for example, shortage of improved seeds is expected until at least 2030 despite the ambitious new seed orchard program (Almqvist et al. 2010).
There is ample evidence that the use of unsuitable seed provenances may markedly reduce adaptation and growth performance of the progenies due to both provenance and maternal environmental effects (Muffler et al. 2016). Variations in environmental conditions within the natural range of the species may favor monomorphic and genetically different populations, polymorphy within populations or clinal variation. For instance, provenance effect appeared to be the most important factor influencing the wood properties of six European populations of P. abies (Sandak et al. 2015). In addition, maternal environmental conditions under which seeds are developed have been proven to have an adverse impact on adaptation and growth of progenies when planted outside their natural environmental conditions. Seedlings raised from seeds reproduced under warm conditions exhibit later flushing, an extended growth period and a delayed development of frost hardiness during early autumn compared with seedlings raised from seeds of the same parents reproduced under colder conditions (Johnsen and Ostreng 1994; Skrøppa et al. 1994; Kohmann and Johnsen 1994; Johnsen et al. 1995, 1996). Although transferring maternal clones to a warmer climate in the south to encourage better floral initiation and seed maturation is a common practice, maternal environment effects are believed to persist for a longer time due to long-lasting epigenetic “memory” regulated by the prevailing temperature and photoperiod during seed production (Besnard et al. 2008).
Thus, monitoring seed provenances of the desired species prior to planting at a given site or region is an important issues to ensure maximum productivity. Seed certification, regulated by seed trade movement legislation, is currently used to monitor seed transfer between countries; however there is no simple and objective method to verify seed movement. Thus, the aim of this study was to evaluate the feasibility of Near Infrared (NIR) spectroscopy as a rapid and non-destructive technique for authenticating P. abies seed provenances. NIR spectroscopy is widely recognized as a rapid technique for quantitative and qualitative analyses of chemical and physical properties in a wide range of organic materials (Workman and Weyer 2012). The NIR spectrum spans from 780–2500 nm wavelength range of the electromagnetic spectrum. It works on the principle that when organic samples interact with NIR radiation, absorption bands are formed as a result of molecular bond vibrations that give rise to overtones and combinations of the fundamental vibrations active in the mid-infrared region (Shenk et al. 2001; Workman and Weyer 2012). The absorption bands typically observed in the NIR region correspond to bonds containing light atoms, such as C–H, O–H, N–H and S–H, which in turn are the major molecular moieties in all biological samples. Previous studies have demonstrated the capability of NIR spectroscopy combined with multivariate modelling for differentiating among sets of similar biological materials, including identification of seed sources and parents of Scots pine (Tigabu et al. 2005), geographical origin of olive oils (Woodcock et al. 2008; Casale et al. 2009; Bevilacqua et al. 2012) and pistachio (Vitale et al. 2013), as well as discrimination of pure and hybrid larch seeds (Farhadi et al. 2016). To our knowledge, there is no study that attempted to apply NIR spectroscopy for authenticating P. abies seed provenances.
Materials and methods
Seed samples
P. abies seed lots were collected from five sites in Sweden, Finland, Poland and Lithuania (Table 1). The seeds were harvested at different time, dried (<10% moisture content) prior to storage and stored at 5 °C. The Swedish and Finnish provenances represented the northern distribution range of the species while the Polish and Lithuanian provenances represented the southern range. From each provenance, a random sample of 750 seeds (n = 150 seeds × 5 seeds lots/provenance) were taken for NIR analysis. To develop the classification model, samples from three seed lots per provenance (n = 450 seeds per provenance) were used as a working sample while samples from the remaining two seed lots (n = 300 seeds per provenance) were assigned as external test to validate the computed model (Table 2).
Spectral acquisition
NIR reflectance spectra, in the form of log (1/R), of individual seeds were acquired by XDS Rapid Content Analyzer (FOSS NIRSystems, Inc.) from 780 to 2498 nm at 0.5 nm resolution. Each individual seed was scanned by directly placing it at the center of the scanning glass window of the instrument with 9 mm aperture. The sample holder was then covered with the instrument’s lid that had a black background with no reflectance, and each individual seed was scanned at stationary position to avoid displacement of the seed during scanning. Prior to collecting single seed reflectance spectra, a reference measurement was taken using the standard built-in reference of the instrument. In addition, reference measurements were taken after every 20 scans to minimize the probable effect of instrumental drift during scanning. For every seed, 32 monochromatic scans were taken and the average value recorded. The spectral data were then exported from Vision Software (FOSS NIRSytems, Inc. VISION 3.5) as NSAS file and imported into Simca-P + software (Version 14.0.0.0, Umetrics AB, Sweden) to develop multivariate classification model.
Multivariate classification modelling
As the first step in multivariate modelling, Principal Component Analysis (PCA) was performed to identify outliers in the data set. Consequently, 71 seeds were found to be outliers and excluded from the final model (Table 2). Particularly, the first 25 seeds from Finland appeared to form clear grouping compared to the rest of the samples. This could be due to measurement error arising from insufficient warm up of the instrument. The final dataset was composed 2929 individual seed spectra, of which 1171 spectra were used for calibration to develop the model. Seed lots included in the calibration set were selected based on their geographic distribution within each country; thereby the model had good representation of intra-provenance variability (Table 2). The robustness of computed classification model was evaluated using internal (n = 600 seeds) and external (n = 1158 seeds) test sets (Table 2). The internal test set was composed of seed samples drawn from the same seed lots used for calibration, but was excluded during model development, whereas seed samples for the external test set were drawn from seed lots which were not included in the model.
NIR spectroscopic data are not usually amenable for direct analysis due to light scattering, base line shift, instrumental drift, and path length differences (Tigabu and Odén 2004; Tigabu et al. 2004), thus such noise in the spectra should be remove using spectral pre-treatment techniques to enhance signal to noise ratio prior to model fitting. In this study, the raw spectra were pre-treated with standard normal variate transformation (SNV) to scale down path-length differences that could arise from variation in individual seed size. The SNV transformation was performed according to the following general formula (Barnes et al. 1989):
where (x ik )* = the transformed absorbance value for the ith object at the kth wavelength, x ik = the original absorbance value for the ith sample at the kth wavelength, m i = the mean of the K spectral measurements for sample i, S i = the standard deviation of the same K measurements and K is the number of X-variables (wavelength channels).
A classification model was then computed with Orthogonal Projection to Latent Structures-Discriminant Analysis (OPLS-DA) of SNV-transformed spectral data set. Generally, the OPLS-DA is an extension to the supervised PLS-DA method featuring an integrated Orthogonal Signal Correction (OSC)-filter (Trygg and Wold 2002). In simple terms, OPLS-DA is a two-step approach where spectral variations that have no correlation with the classes (denoted as Y-orthogonal) were first removed from the spectra, and then discriminant models were fitted on predictive spectral variation. To do this, the OPLS-DA modelling approach uses the information in the categorical response matrix Y (a matrix of dummy variables) to decompose the X matrix (the spectral data) into three distinct parts: (1) the predictive score matrix and its loading matrix for X, (2) the corresponding Y-orthogonal score matrix and loading matrix of Y-orthogonal components, and (3) the residual matrix of X (Bylesjö et al. 2006; Trygg and Wold 2003). Components orthogonal to the response variable containing unwanted systematic variation in the spectra were then subtracted from the original spectral data to produce a filtered descriptor matrix. The final discriminant model was computed using the filtered predictive absorbance values as regressor and a Y-matrix of dummy variables (1 for member of a given class, 0.0 otherwise) as regressand.
The model was fitted on mean-centered data set and the number of significant model components was determined by cross validation. A component was considered significant if the ratio of the prediction error sum of squares (PRESS) to the residual sum of squares of the previous dimension (SS) was statistically smaller than 1.0 (Eriksson et al. 2006). Finally, the computed models were used to classify samples in test sets, and samples were considered as member of a given class if the predicted value was greater than 0.5 and all others were considered as non-members. To evaluate the performance of the classification model, the following classification parameters were computed: sensitivity (Sn), specificity (Sp), classification error rate (ER), Mean classification ER (MER), classification accuracy (CA) and Mean classification accuracy (MCA) following Ballabio and Consonni (2013). The equations used for calculating classification parameters were:
where TP (True Positive) is the number of samples of a given class correctly recognized as member, FN (False Negative) is the number of samples of a given class incorrectly classified as non-member, TN (True Negative) the number of non-member samples correctly classified as non-member of a given class, and FP (False Positive) is the number of non-member samples incorrectly classified as member of a given class, and n is the number of classes. Class sensitivity describes the model’s ability to correctly recognize samples belonging to that class; whereas class specificity describes the model’s ability to reject samples of all other classes. The values for both sensitivity and specificity range from 0 to 1; for example, if none of the samples in a given class was classified as member of other classes (FN = 0), the sensitivity for that class would be equal to 1. Likewise, if none of the non-member samples of a given class was classified as member of that class (FP = 0), the specificity for that class would be equal to 1.
Absorption bands accounted for identification of seed provenances
Absorption bands that accounted for identification P. abies seed provenances were determined using a parameter called Variable Influence on Projection (VIP). The VIP for predictive components (PRED_VIPOPLS) was computed using the following formula (Galindo-Prieto et al. 2015).
where K p is the total number of variables in the model; P is the normalized loadings; a and A p are the number of each predictive component and the total number of predictive components, respectively; SSXcomp and SSYcomp represent the explained sum of squares of ath component for X and Y data matrices, respectively; and SSXcum and SSYcum represent the cumulative explained sum of squares by all A components in the model for X and Y data matrices, respectively. Since the sum of squares of all VIP values is equal to the number of spectral X variables contributed in each calibration model, the average VIP value would be 1 (Wold et al. 1993). Thus, predictors with VIP value greater than 1.0 have a strong influence on the model, but a cut-off around 0.7–0.8 has been suggested to discriminate between relevant and irrelevant predictors (Eriksson et al. 2006).
Results
Spectral profile of seed lots
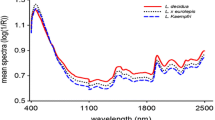
The mean NIR reflectance spectra of seeds from different provenances showed a similar profile with five distinct absorption peaks at 1180, 1514, 1727, 1920, and 2143 nm as well as smaller peaks in the longer NIR region beyond 2100 nm (Fig. 1). Although no unique absorption peak was discerned in the shorter NIR region (780–1100 nm), the absorbance values were high, particularly for Finnish and Swedish seeds. The small jump in the spectra at 1100 nm was caused by a shift in the detection system from Silicon in 780–1100 nm to InGaAs in 1100–2500 nm, but this did affect the OPLS-DA modelling. The subtle difference in absorbance values of seeds from different provenances, particularly between that of Swedish and Finnish, would be difficult to see on the plots at this scale of attenuation. As a whole, it appeared that the NIR spectra contained sufficient information to develop classification models.
Model overview
The four-class model fitted to simultaneously discriminate P. abies seed lots from Sweden, Finland, Lithuania and Poland employed 16.3% of the predictive spectral variation (R2XP) to model 81.1% of the variation among seed provenances in the calibration set (R2Y) with three significant components. The prediction accuracy of the computed classification model (Q2cv), according to cross validation, was 78.2%. The Y-orthogonal spectral variations that were not correlated to the classification of seed provenances (R2Xo) constituted 83.7% of the total spectral variation explained by the model. The 3D score plot for the predictive components showed clear grouping patterns of seed provenances (Fig. 2). The score plots for the first two orthogonal components, which described more than 50% of the variation, revealed that few seed samples irrespective of their provenances were outside the 95% confidence ellipse according to Hotelling’s T2 test—a multivariate generalization of Student’s t test (data not shown). The influence of these samples was, however, filtered during the first step of OPLS-DA modelling approach.
Classification performance
The classification performance of the computed model was evaluated using samples derived from the same seed lots as the calibration (Internal test set) and samples derived from completely different seed lots than the calibration (external test set). For the internal test set, the ability of the model to assign seeds to their respective provenances (sensitivity) as well as its ability to reject seeds of other provenances (specificity) was very high (Table 3). The mean classification accuracy was 99% with mean error rate of 1%. For the external test set, the sensitivity was still high for all seed provenances except Sweden (Table 3) for which sensitivity was relatively low compared to that of the internal test set. However, the specificity was still high for all seed provenances and close to the values obtained for the internal test set. Predicted class membership for external test samples is shown in Fig. 3. For Finnish seed lots, a total of 11 seeds were misclassified, of which 9 seeds were considered Swedish, 1 seed each Lithuanian and Polish. A total of 57 Swedish seeds were misclassified as Finnish. For the Lithuania seed lots, a total of 6 seeds were misclassified of which 3, 2 and 1 seeds were misclassified as Finnish, Polish and Swedish seeds, respectively. For Polish seed lots, a total of 19 samples were misclassified as Lithuanian (12) and Swedish (7). As a whole, the mean classification accuracy was 95% with mean error rate of 5% (Table 3).
Absorption bands accounted for identification of seed provenances
The VIP plot shows that absorption bands in 780–960, 1246–1362, 1889–1980 and 2110–2155 nm, with peaks centered at 850, 1312, 1915 and 2142 nm had a strong influence on identification of seed provenances (VIP > 1; Fig. 4). The wavelength region beyond 2250 nm, with two small peaks at 2355 and 2380 nm, were also highly relevant (VIP > 1.0) for identification of P. abies seed provenances. Other NIR regions of interest that contributed to classification of seed provenances appeared in the 1100–1142 and 1415–1480 nm, with peaks centered at 1122 and 1453 nm (VIP = 0.7–0.8).
Discussion
The results demonstrated that seed provenances of P. abies can be successfully identified by NIR spectroscopy and multivariate modelling. The classification model fitted on SNV-treated data set described 16% of the predictive spectral variation that, in turn, explained 81% of the variation between seed provenances with very good prediction ability according to cross-validation. The model statistics further showed that the computed model was dimensionally less complex (three significant components), which is an important element in the interpretation of multivariate analysis, and parsimonious models with few predictive components are often sought after (Trygg and Wold 2003; Pinto et al. 2012). It is worth noting that NIR spectroscopy is highly sensitive and sufficiently detects subtle differences as low as 0.1% of the total concentration of the analyte (Osborne et al. 1993) while multivariate analysis is more effective in extracting such information from the spectra than univariate analysis (Næs et al. 2002).
The model’s ability to recognize members (sensitivity) while rejecting non-members (specificity) of a given class was excellent for both internal and external test samples (Table 3), which indicates the robustness of the model. Although the specificity of the model for the Swedish samples was excellent (98%), the sensitivity was slightly less for external (81%) than internal (99%) test set samples. This suggests lack of intra-provenance homogeneity of seeds due to variation in size moisture content and chemical composition of individual seeds, which in turn influenced the intensity of recorded signal. As absorbance is a function of concentration from Beer–Lambert’s law, small-sized seeds might have failed to produce a threshold amount of spectral signal well below the mean absorbance values of a particular seed class. Previous studies have also attributed individual seed size variability to differences in path-length difference and light scattering, which in turn influence class discrimination (Farhadi et al. 2015, 2016; Daneshvar et al. 2015; Tigabu and Odén 2004). As a whole, the overall classification accuracy of the classification model is high (95%), with very low error rate (5%). Expanding the range of variability in individual seeds by including more samples into the calibration data set, the sensitivity of the model, particularly for the Swedish samples, could probably be improved.
To get insights into the chemical background of seeds accounted for identification of seed provenances by NIR spectroscopy, the observed absorption bands were interpreted based on previous studies on assignment of bands to functional groups (Osborne et al. 1993; Shenk et al. 2001; Workman and Weyer 2012) and knowledge of reserve compounds in P. abies seeds. In the present study, absorption bands in 780–960, 1246–1362, 1889–1980 and 2110–2155 nm regions were useful in identifying seed provenances (Fig. 4). The absorption band in 780–1100 nm, with a major peak at 850 nm, is characteristic of the third overtone of C–H stretching vibration and second overtone N–H and C–H stretching vibrations. Molecules responsible for absorption in this region are lipid and protein moieties such as CH3, CH2, ArNH2 (aromatic amino acids) and NH2. Previously, high correlation between the amount of fat content in pork and large absorption peak at 928 nm was observed (Norris 1983). The absorption band with a peak at 1312 nm corresponds to C–H combination and first overtone of N–H stretching vibration due to absorption by CH2 and protein moieties.
The dominant peak at 1915 nm arises from O–H stretch/HOH deformation combination and O–H bend second overtone and C=O stretch second overtone due to absorption by several functional groups, notably H2O, –CO2R and starch. Pure water has absorption packneaks at 1940 nm due to O–H stretch first overtone and combination bands involving O–H stretch and O–H bend, although these bands are subject to shifts due to variation in temperature and in hydrogen bonding when water is in a solvent or solute admixture (Osborne et al. 1993). When the 1900–2000 nm spectral range was excluded from the model, the truncated model still identified seed provenances with 90% accuracy. Thus, the dominant absorption peak at 1915 nm found in this study may be attributed to the presence of both seed moisture and starch, as starch grains are abundant in plastids of P. abies seeds before drying for storage (Hakman 1993). The absorption band in 2110–2500 nm is characteristic of CH2 stretch-bend combinations as well as other vibrational modes of molecular bonds. Several fatty acids in oil crops, notably polyunsaturated fatty acids, have shown positive correlation to absorption bands in these regions (Ribeiro et al. 2013; Kim et al. 2007; Hourant et al. 2000). Previous studies have shown good correlations between absorbance values in this spectral region and the content of major fatty acids and can be used as a basis for discrimination of single seeds according to viability class (Farhadi et al. 2015; Daneshvar et al. 2015; Tigabu et al. 2007; Tigabu and Odén 2003).
As a whole, differences in the amount of seed storage reserve compounds, mainly fatty acids and proteins, were the basis for identification of P. abies seed provenances by NIR spectroscopy. Lipids are the dominant reserve compounds in many conifers, including P. abies seeds, which vary between 21.3 and 31.6% with higher amount towards the northern distribution range (Tigabu et al. 2004). Previous studies have also shown that oleic, linoleic and 5,9,12-octadecatrienoic acids are the most abundant fatty acids in the triacylglycerol of P. abies seeds (Tillman-Sutela et al. 1995), and Δ5 unsaturated polymethylene interrupted fatty acids (UPIFAs) constitute 27% of P. abies seeds (Lísa et al. 2007). It was also reported that the total protein content of P. abies seeds varies between 15.7 and 18.7%; being significantly higher for Finnish than Swedish seeds (Tigabu et al. 2004).
In conclusion, the results provide evidence that NIR spectroscopy is a promising method for monitoring putative seed provenances and in seed certification. The technique offers several benefits compared to other methods (e.g. molecular technique), such as high efficiency of sample processing as the seeds don’t have to be prepared for the tests, and it takes ca. 2 min to scan a single seed. In addition, acquiring the spectral data is a simple task and there is possibility for automation of the process. As the method is non-destructive, it is attractive for seed handling and plant-breeding programs where loss of viable seed should be avoided.
References
Almqvist C, Wennström U, Karlsson B (2010) Improved forest regeneration material 2010–2050 Supply and needs, and measures to minimize shortage and maximize genetic gain. Redogörelse nr 3, Skogforsk
Ballabio D, Consonni V (2013) Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal Methods 5:3790–3798
Barnes RJ, Dhanoa MS, Lister SJ (1989) Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl Spectrosc 43:772–777
Besnard G, Acheré V, Jeandroz S, Johnsen Ø, Rampant FP, Baumann R, Müller-Starck G, Skrøppa T, Favre J-M (2008) Does maternal environmental condition during reproductive development induce genotypic selection in Picea abies? Ann For Sci 65:109
Bevilacqua M, Bucci R, Magrì AD, Magrì AL, Marini F (2012) Tracing the origin of extra virgin olive oils by infrared spectroscopy and chemometrics: a case study. Anal Chim Acta 717:39–51
Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometr 20:341–351
Casale M, Casolino C, Oliveri P, Forina M (2009) The potential of coupling information using three analytical techniques for identifying the geographical origin of Liguria extra virgin olive oil. Food Chem 118:163–170
Daneshvar A, Tigabu M, Karimidoost A, Odén PC (2015) Single seed near infrared spectroscopy discriminates viable and non-viable seeds of Juniperus polycarpos. Silva Fenn 49:14. Article ID 1334
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S (2006) Multi- and megavariate data analysis. Basic principles and applications. Second revised and enlarged edition. Umetrics Academy, Umeå
Farhadi M, Tigabu M, Odén PC (2015) Near Infrared Spectroscopy as non-destructive method for sorting viable, petrified and empty seeds of Larix sibirica. Silva Fenn 49:12. Article ID 1340
Farhadi M, Tigabu M, Stener L-G, Odén PC (2016) Feasibility of Vis + NIR spectroscopy for non-destructive verification of European × Japanese larch hybrid seeds. New For 47(2):271–285
Galindo-Prieto B, Eriksson L, Trygg J (2015) Variable influence on projection (VIP) for OPLS models and its applicability in multivariate time series analysis. Chemom Intell Lab Syst 146:297–304
Ge ZM, Kellomäki S, Peltola H, Zhou X, Wang KY, Väisänen H (2011) Impacts of changing climate on the productivity of Norway spruce dominant stands with a mixture of Scots pine and birch in relation to water availability in southern and northern Finland. Tree Physiol 31:323–338
Hakman I (1993) Embryology in Norway spruce (Picea abies). An analysis of the composition of seed storage proteins and deposition of storage reserves during seed development and somatic embryogenesis. Physiol Plant 87:148–159
Hourant P, Baeten V, Morales MT, Meurens M, Aparicio R (2000) Oil and fat classification by selected bands of near-infrared spectroscopy. Appl Spectrosc 54:1168–1174
Johnsen Ø, Ostreng G (1994) Effects of plus tree selection and seed orchard environment on progenies of Picea abies. Can J For Res 24:32–38
Johnsen Ø, Skrøppa T, Haug G, Apeland I, Ostreng G (1995) Sexual reproduction in a greenhouse and reduced autumn frost-hardiness of Picea abies progenies. Tree Physiol 15:551–555
Johnsen Ø, Skrøppa T, Junttila O, Dæhlen OG (1996) Influence of the female flowering environment on autumn frost-hardiness of Picea abies progenies. Theor Appl Genet 92:797–802
Kim KS, Park SH, Choung MG, Jang YS (2007) Use of Near-infrared spectroscopy for estimating fatty acid composition in intact seeds of rapeseed. J Crop Sci Biotech 10:15–20
Kohmann K, Johnsen Ø (1994) The timing of bud-set in seedlings of Picea abies from seed crops of a cool versus a warm summer. Silvae Genet 43:328–332
Koprowski M, Zielski A (2006) Dendrochronology of Norway spruce (Picea abies (L.) Karst.) from two range centres in lowland Poland. Trees 20:383–390
Lísa M, Holčapek M, Řezanka T, Kabátová N (2007) High-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry and gas chromatography-flame ionization detection characterization of Δ5-polyenoic fatty acids in triacylglycerols from conifer seed oils. J Chromatogr A 1146:67–77
Muffler L, Beierkuhnlein C, Aas G, Jentsch A, Schweiger AH, Zohner C, Kreyling J (2016) Distribution ranges and spring phenology explain late frost sensitivity in 170 woody plants from the Northern Hemisphere. Glob Ecol Biogeogr 25:1061–1071
Næs T, Isaksson T, Fearn T, Davies T (2002) A user friendly guide to multivariate calibration and classification. NIR Publications, Chichester
Norris KH (1983) Multivariate analysis of raw materials. In: Shemilt LW (ed) Chemistry and world food supplies: the new frontiers. Pergamon press, New York, pp 527–535
Norwegian Ministry of the Environment (2009) Norway`s fifth national communication under the framework convention on climate change. Norwegian Government Administration Services, Oslo
Osborne BG, Fearn T, Hindle PH (1993) Practical NIR spectroscopy with applications in food and beverage analysis. Longman, Harlow
Pinto RC, Trygg J, Gottfries J (2012) Advantages of orthogonal inspection in chemometrics. J Chemom 26:231–235
Ribeiro LF, Peralta-Zamora PG, Maia BHLNS, Ramos LP, Pereira-Netto AB (2013) Prediction of linolenic and linoleic fatty acids content in flax seeds and flax seeds flours through the use of infrared reflectance spectroscopy and multivariate calibration. Food Res Int 51:848–854
Sandak J, Sandak A, Cantini C, Autino A (2015) Differences in wood properties of Picea abies L. Karst. in relation to site of provenance and population genetics. Holzforschung 69(4):385–397
Shenk JS, Workman JJ, Westerhaus MO (2001) Application of NIR spectroscopy to agricultural products. In: Burnes DA, Ciurczak EW (eds) Handbook of near-infrared analysis. Marcel Dekker, New York, pp 419–474
Skrøppa T, Nikkanen T, Routsalainen S, Johnsen Ø (1994) Effects of sexual reproduction at different latitudes on performance of the progeny of Picea abies. Silvae Genet 43:297–303
Statistical Yearbook of Forestry (2011) Official statistics of Sweden. National Board of Forestry, Jönköping
Tigabu M, Odén PC (2003) Classification of viable and empty seeds of Pinus patula Schiede & Deppe with near-infrared spectroscopy and multivariate analysis. New For 25:163–176
Tigabu M, Odén PC (2004) Simultaneous detection of filled, empty and insect-infested seeds of three Larix species with single seed near-infrared transmittance spectroscopy. New For 27:39–53
Tigabu M, Odén PC, Shen TY (2004) Application of near infrared spectroscopy for the detection of internal insect infestation in Picea abies seed lots. Can J For Res 34:76–84
Tigabu M, Odén PC, Lindgren D (2005) Identification of seed sources and parents of Pinus sylvestris L. using visible–near infrared reflectance spectra and multivariate analysis. Trees 19:468–476
Tigabu M, Fjellström J, Odén PC, Teketay D (2007) Germination of Juniperus procera seeds in response to stratification and smoke treatments, and detection of insect-damaged seeds with VIS + NIR spectroscopy. New For 33:155–169
Tillman-Sutela E, Johansson A, Laasko P, Mattila T, Kallio H (1995) Triacylglycerols in the seeds of northern Scots pine, Pinus sylvesteris and Norway spruce, Picea abies (L.) Karst. Trees 10:40–45
Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemom 16:119–128
Trygg J, Wold S (2003) O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J Chemom 17:53–64
Vitale R, Bevilacqua M, Bucci R, Magrì AD, Magrì AL, Marini F (2013) A rapid and non-invasive method for authenticating the origin of pistachio samples by NIR spectroscopy and chemometrics. Chemometr Intell Lab Syst 121:90–99
Vitas A (2004) Tree rings of Norway spruce (Picea abies (L.) Karst) in Lithuania as drought indicators: dendroecological approach. Pol J Ecol 52:201–210
Wold S, Johansson E, Cocchi M (1993) PLS-partial least squares projections to latent structures. In: Kubinyi H (ed) 3D QSAR in drug design: theory, methods and applications. ESCOM, Leiden, pp 523–550
Woodcock T, Downey G, O’Donnell C (2008) Confirmation of declared provenance of European extra virgin olive oil samples by NIR spectroscopy. J Agric Food Chem 56:11520–11525
Workman J, Weyer L (2012) Practical guide and spectral atlas for interpretive near-infrared spectroscopy, 2nd edn. CRC Press, Boca Raton
Acknowledgements
The study was financially supported by Karl Erik Önnesjös Foundation. We thank the following institutions and persons for providing us with seed samples and associated information: Skogforsk in Sävar, Sweden (Monica Lundström), Finnish Food Safety Authority Evira, Plant Health Unit/Forest Reproductive Material (Dr. Kari Leinonen and Metti Salminen), Polish State Forest Department, RDLP in Krakow (Dr. Robert Głodowski) and Faculty of Forestry AUC (Dr. Jacek Banach) and Lithuanian Forest Districts. Our special thanks go to Dr. Vilis Brukas at SLU for assisting in networking. We extend our gratitude to the associate editor and two anonymous reviewers for valuable and constructive comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Farhadi, M., Tigabu, M., Pietrzykowski, M. et al. Application of near infrared spectroscopy for authentication of Picea abies seed provenance. New Forests 48, 629–642 (2017). https://doi.org/10.1007/s11056-017-9589-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-017-9589-1