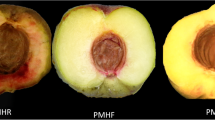
Abstract
Many Litchi chinensis cv. Baitangying orchards are suffering from a serious fruit cracking problem, but few studies have improved our understanding of the mechanism or the molecular basis of cracking susceptibility in ‘Baitangying’. We conducted metabolome and transcriptome analyses of three types of litchi pericarps. To prevent passive progression after fruit cracking from affecting the results, we mainly focused on 11 metabolites and 101 genes that showed the same regulatory status and overlap in pairwise comparisons of cracking ‘Baitangying’ versus noncracking ‘Baitangying’ and noncracking ‘Baitangying’ versus noncracking ‘Feizixiao’. Compared with the cracking-resistant cultivar ‘Feizixiao’, the ‘Baitangying’ pericarp has higher abscisic acid contents, and the presence of relevant metabolites and genes suggests increased biosynthesis of ethylene and jasmonic acid and decreased auxin and brassinosteroid biosynthesis. The fruit cracking-susceptible trait in ‘Baitangying’ might be associated with differences in the balance of these five types of hormones between the pericarp of this cultivar and that of ‘Feizixiao’. Additionally, combined analyses showed a correspondence between the metabolite profiles and transcript patterns. qRT-PCR validation indicated the reliability of our high-throughput results. The acquired information might help in further studying the mechanisms that mediate fruit cracking susceptibility in ‘Baitangying’ and other litchi cultivars.






Similar content being viewed by others
References
Khadivi-Khub A (2014) Physiological and genetic factors influencing fruit cracking. Acta Physiol Plant 37(1):1718. https://doi.org/10.1007/s11738-014-1718-2
Tate D (2014) Tropical fruit. Archipelago Press, Brooklyn
Jiang S-Y, Xu H-Y, Wang H-C, Hu G-b, Li J-G, Chen H-B, Huang X-m (2012) A comparison of the costs of flowering in ‘Feizixiao’ and ‘Baitangying’ litchi. Sci Hortic 148:118–125. https://doi.org/10.1016/j.scienta.2012.09.035
Li W-C, Wu J-Y, Zhang H-N, Shi S-Y, Liu L-Q, Shu B, Liang Q-Z, Xie J-H, Wei Y-Z (2014) De novo assembly and characterization of pericarp transcriptome and identification of candidate genes mediating fruit cracking in Litchi chinensis Sonn. Int J Mol Sci 15(10):17667–17685. https://doi.org/10.3390/ijms151017667
Li JG, Huang HB, Gao FF, Huang XM, Wang HC (2001) An overview of litchi fruit cracking. Acta Hortic 558:205–208. https://doi.org/10.17660/ActaHortic.2001.558.28
Wang Y, Lu W, Li J, Jiang Y (2006) Differential expression of two expansin genes in developing fruit of cracking-susceptible and -resistant litchi cultivars. J Am Soc Hortic Sci 131(1):118–121
Wang J, Gao X, Ma Z, Chen J, Liu Y (2019) Analysis of the molecular basis of fruit cracking susceptibility in Litchi chinensis cv. Baitangying by transcriptome and quantitative proteome profiling. J Plant Physiol 234–235:106–116. https://doi.org/10.1016/j.jplph.2019.01.014
Will S, Eichert T, Fernández V, Müller T, Römheld V (2012) Boron foliar fertilization of soybean and lychee: effects of side of application and formulation adjuvants. J Plant Nut Soil Sci 175(2):180–188. https://doi.org/10.1002/jpln.201100107
Li C, Wang Y, Ying P, Ma W, Li J (2015) Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front Plant Sci 6:502. https://doi.org/10.3389/fpls.2015.00502
Li C, Wang Y, Huang X, Li J, Wang H, Li J (2015) An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front Plant Sci 6:439. https://doi.org/10.3389/fpls.2015.00439
Lai B, Hu B, Qin Y-H, Zhao J-T, Wang H-C, Hu G-B (2015) Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genomics 16(1):225. https://doi.org/10.1186/s12864-015-1433-4
Yun Z, Qu H, Wang H, Zhu F, Zhang Z, Duan X, Yang B, Cheng Y, Jiang Y (2016) Comparative transcriptome and metabolome provides new insights into the regulatory mechanisms of accelerated senescence in litchi fruit after cold storage. Sci Rep 6:19356. https://doi.org/10.1038/srep19356
Luhua S, Hegie A, Suzuki N, Shulaev E, Luo X, Cenariu D, Ma V, Kao S, Lim J, Gunay MB, Oosumi T, Lee SC, Harper J, Cushman J, Gollery M, Girke T, Bailey-Serres J, Stevenson RA, Zhu J-K, Mittler R (2013) Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol Plant 148(3):322–333. https://doi.org/10.1111/ppl.12013
Zhou G, Wang X, Yan F, Wang X, Li R, Cheng J, Lou Y (2011) Genome-wide transcriptional changes and defence-related chemical profiling of rice in response to infestation by the rice striped stem borer Chilo suppressalis. Physiol Plant 143(1):21–40. https://doi.org/10.1111/j.1399-3054.2011.01483.x
Eisenmann P, Ehlers M, Weinert C, Tzvetkova P, Silber M, Rist M, Luy B, Muhle-Goll C (2016) Untargeted NMR spectroscopic analysis of the metabolic variety of new apple cultivars. Metabolites 6(3):29
Lin Q, Wang C, Dong W, Jiang Q, Wang D, Li S, Chen M, Liu C, Sun C, Chen K (2015) Transcriptome and metabolome analyses of sugar and organic acid metabolism in Ponkan (Citrus reticulata) fruit during fruit maturation. Gene 554(1):64–74. https://doi.org/10.1016/j.gene.2014.10.025
Ibáñez AM, Martinelli F, Reagan RL, Uratsu SL, Vo A, Tinoco MA, Phu ML, Chen Y, Rocke DM, Dandekar AM (2014) Transcriptome and metabolome analysis of Citrus fruit to elucidate puffing disorder. Plant Sci 217–218:87–98. https://doi.org/10.1016/j.plantsci.2013.12.003
Domingos S, Fino J, Paulo OS, Oliveira CM, Goulao LF (2016) Molecular candidates for early-stage flower-to-fruit transition in stenospermocarpic table grape (Vitis vinifera L.) inflorescences ascribed by differential transcriptome and metabolome profiles. Plant Sci 244:40–56. https://doi.org/10.1016/j.plantsci.2015.12.009
Khalil MNA, Fekry MI, Farag MA (2017) Metabolome based volatiles profiling in 13 date palm fruit varieties from Egypt via SPME GC–MS and chemometrics. Food Chem 217:171–181. https://doi.org/10.1016/j.foodchem.2016.08.089
Sobolev AP, Neelam A, Fatima T, Shukla V, Handa AK, Mattoo AK (2014) Genetic introgression of ethylene-suppressed transgenic tomatoes with higher-polyamines trait overcomes many unintended effects due to reduced ethylene on the primary metabolome. Front Plant Sci 5:632. https://doi.org/10.3389/fpls.2014.00632
Huang XM, Wang HC, Gao FF, Huang HB (1999) A comparative study of the pericarp of litchi cultivars susceptible and resistant to fruit cracking. J Hortic Sci Biotechnol 74(3):351–354. https://doi.org/10.1080/14620316.1999.11511120
Jing J, Shi Y, Zhang Q, Wang J, Ruan J (2017) Prediction of Chinese green tea ranking by metabolite profiling using ultra-performance liquid chromatography–quadrupole time-of-flight mass spectrometry (UPLC–Q-TOF/MS). Food Chem 221:311–316. https://doi.org/10.1016/j.foodchem.2016.10.068
Zhou X, Liu L, Lan X, Cohen D, Zhang Y, Ravindran AV, Yuan S, Zheng P, Coghill D, Yang L, Hetrick SE, Jiang X, Benoliel J-J, Cipriani A, Xie P (2018) Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. https://doi.org/10.1038/s41380-018-0047-z
Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78(3):779–787. https://doi.org/10.1021/ac051437y
Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J (2008) Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem 80(1):115–122. https://doi.org/10.1021/ac0713510
Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemom 16(3):119–128. https://doi.org/10.1002/cem.695
Haynes W (2013) Student’s t-test. Encyclopedia of Systems Biology. Springer, New york, pp 2023–2025
Patel RK, Jain M (2015) NGS QC Toolkit: a platform for quality control of next-generation sequencing data. In: Nelson KE (ed) Encyclopedia of metagenomics: genes, genomes and metagenomes: basics, methods, databases and tools. Springer, Boston, pp 544–548. https://doi.org/10.1007/978-1-4899-7478-5_348
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494. https://doi.org/10.1038/nprot.2013.084
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10(1):421. https://doi.org/10.1186/1471-2105-10-421
The UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43(D1):D204–D212. https://doi.org/10.1093/nar/gku989
Wu J, Mao X, Cai T, Luo J, Wei L (2006) KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 34(suppl_2):W720–W724. https://doi.org/10.1093/nar/gkl167
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621. https://doi.org/10.1038/nmeth.1226
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12(1):323. https://doi.org/10.1186/1471-2105-12-323
Zhou X, Lindsay H, Robinson MD (2014) Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 42(11):e91. https://doi.org/10.1093/nar/gku310
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B 57(1):289–300
Zhan C, Li X, Zhao Z, Yang T, Wang X, Luo B, Zhang Q, Hu Y, Hu X (2016) Comprehensive analysis of the triterpenoid saponins biosynthetic pathway in Anemone flaccida by transcriptome and proteome profiling. Front Plant Sci 7:1094. https://doi.org/10.3389/fpls.2016.01094
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Tarachiwin L, Ute K, Kobayashi A, Fukusaki E (2007) 1H NMR based metabolic profiling in the evaluation of japanese green tea quality. J Agric Food Chem 55(23):9330–9336. https://doi.org/10.1021/jf071956x
Shen Q, Fu L, Dai F, Jiang L, Zhang G, Wu D (2016) Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genom 17(1):889. https://doi.org/10.1186/s12864-016-3242-9
Sade D, Shriki O, Cuadros-Inostroza A, Tohge T, Semel Y, Haviv Y, Willmitzer L, Fernie AR, Czosnek H, Brotman Y (2015) Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 11(1):81–97. https://doi.org/10.1007/s11306-014-0670-x
Lu X, Kim H, Zhong S, Chen H, Hu Z, Zhou B (2014) De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC Genomics 15(1):805. https://doi.org/10.1186/1471-2164-15-805
Pathak AK, Singh SP, Gupta Y, Gurjar AKS, Mantri SS, Tuli R (2016) Transcriptional changes during ovule development in two genotypes of litchi (Litchi chinensis Sonn) with contrast in seed size. Sci Rep 6:36304. https://doi.org/10.1038/srep36304
Domínguez E, Cuartero J, Heredia A (2011) An overview on plant cuticle biomechanics. Plant Sci 181(2):77–84. https://doi.org/10.1016/j.plantsci.2011.04.016
Douglas CJ (1996) Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci 1(6):171–178. https://doi.org/10.1016/1360-1385(96)10019-4
Marmon SK, Sturtevant D, Herrfurth C, Chapman KD, Stymne S, Feussner I (2017) Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol 173(4):2081. https://doi.org/10.1104/pp.16.01865
Yang W, Pollard M, Li-Beisson Y, Ohlrogge J (2016) Quantitative analysis of glycerol in dicarboxylic acid-rich cutins provides insights into Arabidopsis cutin structure. Phytochemistry 130:159–169. https://doi.org/10.1016/j.phytochem.2016.03.017
Petit J, Bres C, Mauxion J-P, Wong Jun Tai F, Martin LBB, Fich EA, Joubès J, Rose JKC, Domergue F, Rothan C (2016) The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol 171(2):894–913. https://doi.org/10.1104/pp.16.00409
Balbontín C, Ayala H, Rubilar J, Cote J, Figueroa CR (2014) Transcriptional analysis of cell wall and cuticle related genes during fruit development of two sweet cherry cultivars with contrasting levels of cracking tolerance. Chil J Agric Res 74(2):162–169. https://doi.org/10.4067/S0718-58392014000200006
Berger A, Kostyan MK, Klose SI, Gastegger M, Lorbeer E, Brecker L, Schinnerl J (2015) Loganin and secologanin derived tryptamine–iridoid alkaloids from Palicourea crocea and Palicourea padifolia (Rubiaceae). Phytochemistry 116:162–169. https://doi.org/10.1016/j.phytochem.2015.05.013
Bais HP, Walker TS, Stermitz FR, Hufbauer RA, Vivanco JM (2002) Enantiomeric-dependent phytotoxic and antimicrobial activity of (±)-Catechin. A rhizosecreted racemic mixture from spotted knapweed. Plant Physiol 128(4):1173–1179. https://doi.org/10.1104/pp.011019
Chen X, Chen D, Jiang H, Sun H, Zhang C, Zhao H, Li X, Yan F, Chen C, Xu Z (2018) Aroma characterization of Hanzhong black tea (Camellia sinensis) using solid phase extraction coupled with gas chromatography–mass spectrometry and olfactometry and sensory analysis. Food Chem 274:130–136. https://doi.org/10.1016/j.foodchem.2018.08.124
del Mar Caja M, Preston C, Kempf M, Schreier P (2007) Flavor authentication studies of α-Ionone, β-Ionone, and α-Ionol from various sources. J Agric Food Chem 55(16):6700–6704. https://doi.org/10.1021/jf070805r
Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40(D1):D1202–D1210. https://doi.org/10.1093/nar/gkr1090
Shu K, Liu X-d, Xie Q, He Z-h (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9(1):34–45. https://doi.org/10.1016/j.molp.2015.08.010
Strader LC, Zhao Y (2016) Auxin perception and downstream events. Curr Opin Plant Biol 33:8–14. https://doi.org/10.1016/j.pbi.2016.04.004
Arnao MB, Hernández-Ruiz J (2014) Melatonin: plant growth regulator and/or biostimulator during stress? Trends Plant Sci 19(12):789–797. https://doi.org/10.1016/j.tplants.2014.07.006
Hagen G (2015) Auxin signal transduction. Essays Biochem 58:1–12. https://doi.org/10.1042/bse0580001
Sun X, Li Y, He W, Ji C, Xia P, Wang Y, Du S, Li H, Raikhel N, Xiao J, Guo H (2017) Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nat Commun 8:15758. https://doi.org/10.1038/ncomms15758
Wang S, Saito T, Ohkawa K, Ohara H, Suktawee S, Ikeura H, Kondo S (2018) Abscisic acid is involved in aromatic ester biosynthesis related with ethylene in green apples. J Plant Physiol 221:85–93. https://doi.org/10.1016/j.jplph.2017.12.007
Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53(377):2039–2055. https://doi.org/10.1093/jxb/erf072
Tattersall DB, van Heeswijck R, Hoj PB (1997) Identification and characterization of a fruit-specific, thaumatin-like protein that accumulates at very high levels in conjunction with the onset of sugar accumulation and berry softening in grapes. Plant Physiol 114(3):759–769. https://doi.org/10.1104/pp.114.3.759
Prasanna V, Prabha TN, Tharanathan RN (2007) Fruit ripening phenomena–An overview. Crit Rev Food Sci 47(1):1–19. https://doi.org/10.1080/10408390600976841
Vukašinović N, Russinova E (2018) BRexit: possible brassinosteroid export and transport routes. Trends Plant Sci 23(4):285–292. https://doi.org/10.1016/j.tplants.2018.01.005
Koo AJ (2018) Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochem Rev 17(1):51–80. https://doi.org/10.1007/s11101-017-9510-8
Yilmaz C, Ozguven AI (2006) Hormone physiology of preharvest fruit cracking in pomegranate (Punica granatum L.). Acta Hortic 727:545–550. https://doi.org/10.17660/ActaHortic.2006.727.67
Giné-Bordonaba J, Echeverria G, Ubach D, Aguiló-Aguayo I, López ML, Larrigaudière C (2017) Biochemical and physiological changes during fruit development and ripening of two sweet cherry varieties with different levels of cracking tolerance. Plant Physiol Biochem 111:216–225. https://doi.org/10.1016/j.plaphy.2016.12.002
Rudell DR, Fellman JK, Mattheis JP (2005) Preharvest application of methyl jasmonate to ‘Fuji’ apples enhances red coloration and affects fruit size, splitting, and bitter pit incidence. HortScience 40(6):1760–1762
Acknowledgements
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630062016005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2019_4986_MOESM1_ESM.tif
Supplementary material 1 (TIFF 444 kb) Fig. S1 Permutation test (n = 200) of the orthogonal projections to latent structures-discriminant analysis (OPLS-DA) models for the comparative analysis of the three groups (CB vs. B, B vs. F, and CB vs. F) in the positive (above the dotted line) and negative (below the dotted line) detection modes. The green dots and the blue squares represent the R2 and Q2 values, respectively, obtained by the permutation test, and the two dashed lines represent their regression lines. B, ‘Baitangying’; CB, cracking ‘Baitangying’; F, ‘Feizixiao’; R2, coefficient; Q2, cross-validated correlation coefficient
11033_2019_4986_MOESM2_ESM.tif
Supplementary material 2 (TIFF 1607 kb) Fig. S2 Volcano plots of the differentially expressed metabolites (DEM) identified in the three comparative analyses: CB vs. B (A), B vs. F (B), and CB vs. F (C). The bubble size represents the VIP value of each potential biomarker. The red and blue bubbles represent the upregulated and downregulated biomarkers, respectively, and the gray bubble indicates that the biomarker was not found to be significantly differentially expressed in the comparative analysis
11033_2019_4986_MOESM5_ESM.xlsx
Supplementary material 5 (XLSX 28 kb) Table S3 Differentially expressed metabolites (DEMs) identified in the three comparative analyses: CB vs. B (Sheet 1), B vs. F (Sheet 2), and CB vs. F (Sheet 3). The VIP value is the variable importance in the projection of the first principal component in orthogonal projections to latent structures-discriminant analysis (OPLS-DA) model, the Q value is the corrected P value in Student’s t-test, and the fold change was calculated as the ratio of the case_relative content to the control_relative content
11033_2019_4986_MOESM6_ESM.xlsx
Supplementary material 6 (XLSX 28 kb) Table S4 Continuously up/downregulated transcripts from the CB to F pericarps. Detailed information of these transcripts, including sequencing ID, annotation (predicted function, gene name, UniProt ID, GO ID and KEGG pathway), the log2 (fold change) and FDR values obtained in two comparative analyses (CB vs. B and B vs. F), and continuously regulated status (down/up), is shown
Rights and permissions
About this article
Cite this article
Wang, JG., Gao, XM., Ma, ZL. et al. Metabolomic and transcriptomic profiling of three types of litchi pericarps reveals that changes in the hormone balance constitute the molecular basis of the fruit cracking susceptibility of Litchi chinensis cv. Baitangying. Mol Biol Rep 46, 5295–5308 (2019). https://doi.org/10.1007/s11033-019-04986-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04986-2