Abstract
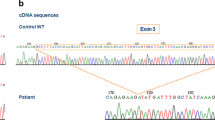
Chronic granulomatous disease (CGD) is a prototypical inborn error of immunity affecting phagocytes, in which these cells are unable to produce reactive oxygen species. CGD is caused by defects in genes encoding subunits of the NADPH oxidase enzyme complex (CYBA, CYBB, CYBC1, NCF1, NCF2, NCF4); inflammatory responses are dysregulated, and patients are highly susceptible to recurrent severe bacterial and fungal infections. X-linked CGD (XL-CGD), caused by mutations in the CYBB gene, is the most common and severe form of CGD. In this study, we describe the analytical processes undertaken in 3 families affected with XL-CGD to illustrate several molecular challenges in the genetic diagnosis of this condition: in family 1, a girl with a heterozygous deletion of CYBB exon 13 and skewed X-chromosome inactivation (XCI); in family 2, a boy with a hemizygous deletion of CYBB exon 7, defining its consequences at the mRNA level; and in family 3, 2 boys with the same novel intronic variant in CYBB (c.1151 + 6 T > A). The variant affected the splicing process, although a small fraction of wild-type mRNA was produced. Their mother was a heterozygous carrier, while their maternal grandmother was a carrier in form of gonosomal mosaicism. In summary, using a variety of techniques, including an NGS-based targeted gene panel and deep amplicon sequencing, copy number variation calling strategies, microarray-based comparative genomic hybridization, and cDNA analysis to define splicing defects and skewed XCI, we show how to face and solve some uncommon genetic mechanisms in the diagnosis of XL-CGD.




Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
van den Berg JM, van Koppen E, Ahlin A, Belohradsky BH, Bernatowska E, Corbeel L, et al. Chronic granulomatous disease: the European experience. PLoS ONE. 2009;4:e5234.
Slack MA, Thomsen IP. Prevention of infectious complications in patients with chronic granulomatous disease. J Pediatric Infect Dis Soc. 2018;7:S25-30.
Kohn DB, Booth C, Kang EM, Pai S-Y, Shaw KL, Santilli G, et al. Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat Med. 2020;26:200–6.
Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human inborn errors of immunity: 2022 update on the classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42:1473–507.
Yu H-H, Yang Y-H, Chiang B-L. Chronic granulomatous disease: a comprehensive review. Clin Rev Allergy Immunol. 2021;61:101–13.
Roos D, de Boer M. Molecular diagnosis of chronic granulomatous disease. Clin Exp Immunol. 2014;175:139–49.
Dinauer MC, Orkin SH, Brown R, Jesaitis AJ, Parkos CA. The glycoprotein encoded by the X-linked chronic granulomatous disease locus is a component of the neutrophil cytochrome b complex. Nature. 1987;327:717–20.
Roos D, van Leeuwen K, Hsu AP, Priel DL, Begtrup A, Brandon R, et al. Hematologically important mutations: X-linked chronic granulomatous disease (fourth update). Blood Cells Mol Dis. 2021;90:102587.
Meischl C, Boer M, Ahlin A, Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet. 2000;8:697–703.
Yu L, Li W, Lv G, Sun G, Yang L, Chen J, et al. De novo somatic mosaicism of CYBB caused by intronic LINE-1 element insertion resulting in chronic granulomatous disease. J Clin Immunol. 2023;43:88–100.
Shvetsova E, Sofronova A, Monajemi R, Gagalova K, Draisma HHM, White SJ, et al. Skewed X-inactivation is common in the general female population. Eur J Hum Genet. 2019;27:455–65.
Gavrilova T, Zelig A, Lee DH. Variable presentation of the CYBB mutation in one family, approach to management, and a review of the literature. Case Rep Med. 2020;2020:2546190.
Arnadottir GA, Norddahl GL, Gudmundsdottir S, Agustsdottir AB, Sigurdsson S, Jensson BO, et al. A homozygous loss-of-function mutation leading to CYBC1 deficiency causes chronic granulomatous disease. Nat Commun. 2018;9:4447.
Kulkarni M, Hule G, de Boer M, van Leeuwen K, Kambli P, Aluri J, et al. Approach to molecular diagnosis of chronic granulomatous disease (CGD): an experience from a large cohort of 90 Indian patients. J Clin Immunol. 2018;38:898–916.
Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65.
Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–83.
Duan J, Zhang J-G, Deng H-W, Wang Y-P. Comparative studies of copy number variation detection methods for next-generation sequencing technologies. PLoS ONE. 2013;8:e59128.
Moreno-Cabrera JM, Del Valle J, Castellanos E, Feliubadaló L, Pineda M, Brunet J, et al. Evaluation of CNV detection tools for NGS panel data in genetic diagnostics. Eur J Hum Genet. 2020;28:1645–55.
Battersby AC, Cale AM, Goldblatt D, Gennery AR. Clinical manifestations of disease in X-linked carriers of chronic granulomatous disease. J Clin Immunol. 2013;33:1276–84.
Pereira G, Dória S. X-chromosome inactivation: implications in human disease. J Genet. 2021;100:63.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–39.
Sauteraud R, Stahl JM, James J, Englebright M, Chen F, Zhan X, et al. Inferring genes that escape X-Chromosome inactivation reveals important contribution of variable escape genes to sex-biased diseases. Genome Res. 2021;31:1629–37.
Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136:1093–111.
Alsina L, Colobran R, de Sevilla MF, Català A, Viñas L, Ricart S, et al. Novel and atypical splicing mutation in a compound heterozygous UNC13D defect presenting in familial hemophagocytic lymphohistiocytosis triggered by EBV infection. Clin Immunol. 2014;153:292–7.
Zhang P, Philippot Q, Ren W, Lei W-T, Li J, Stenson PD, et al. Genome-wide detection of human variants that disrupt intronic branchpoints. Proc Natl Acad Sci U S A. 2022;119:e2211194119.
Mensa-Vilaró A, Bravo García-Morato M, de la Calle-Martin O, Franco-Jarava C, Martínez-Saavedra MT, González-Granado LI, et al. Unexpected relevant role of gene mosaicism in patients with primary immunodeficiency diseases. J Allergy Clin Immunol. 2019;143:359–68.
Aluri J, Cooper MA. Genetic mosaicism as a cause of inborn errors of immunity. J Clin Immunol. 2021;41:718–28.
Batlle-Masó L, Garcia-Prat M, Parra-Martínez A, Franco-Jarava C, Aguiló-Cucurull A, Velasco P, et al. Detection and evolutionary dynamics of somatic FAS variants in autoimmune lymphoproliferative syndrome: diagnostic implications. Front Immunol. 2022;13:1014984.
Labrosse R, Abou-Diab J, Blincoe A, Cros G, Luu TM, Deslandres C, et al. Very early-onset inflammatory manifestations of X-linked chronic granulomatous disease. Front Immunol. 2017;8:1167.
Claps A, Della Corte M, GerocarniNappo S, Francalanci P, Palma P, Finocchi A. How should eosinophilic cystitis be treated in patients with chronic granulomatous disease? Pediatr Nephrol. 2014;29:2229–33.
Acknowledgements
We are deeply grateful to the affected individuals who participated in this study and their families. We acknowledge Celine Cavallo for extensive revision of language and grammar. This research is supported by the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases (ERN-RITA).
Funding
This study was funded by Instituto de Salud Carlos III, grants PI18/00346 and PI20/00761, co-financed by the European Regional Development Fund (ERDF), and by the Jeffrey Modell Foundation. Laura Batlle-Masó has received research support from European Union-NextGenerationEU, Ministry of Universities and Recovery, Transformation and Resilience Plan, through a call from Pompeu Fabra University (Barcelona).
Author information
Authors and Affiliations
Contributions
LBM, PSP, and RC were responsible for the study design. JGR, AMN, and PSP provided patient care, and collected and provided clinical data. AAC performed sample collection and Sanger sequencing. MMG was responsible for flow cytometry studies. CFJ, MGP, and APM performed laboratory analyses and molecular studies. NC was responsible for microarray-based comparative genomic hybridization data. LBM and RC analyzed and interpreted the data, and wrote the manuscript. PSP and RC designed and supervised the project, provided resources, and edited the manuscript. All co-authors reviewed, commented on, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Vall d’Hebron University Hospital [date: 11/06/2021 / code: PR(AG)202/2021].
Consent to Participate
Informed consent was obtained from all individual participants included in the study. For pediatric patients (under 18 years old), written informed consent was obtained from the parents.
Consent for Publication
The authors affirm that human research participants provided informed consent for publication of data contained in this manuscript.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pere Soler-Palacín and Roger Colobran are senior corresponding authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Batlle-Masó, L., Rivière, J.G., Franco-Jarava, C. et al. Molecular Challenges in the Diagnosis of X-Linked Chronic Granulomatous Disease: CNVs, Intronic Variants, Skewed X-Chromosome Inactivation, and Gonosomal Mosaicism. J Clin Immunol 43, 1953–1963 (2023). https://doi.org/10.1007/s10875-023-01556-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-023-01556-x