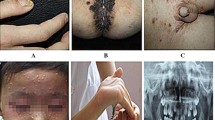
Abstract
Rare, biallelic loss-of-function mutations in DOCK8 result in a combined immune deficiency characterized by severe and recurrent cutaneous infections, eczema, allergies, and susceptibility to malignancy, as well as impaired humoral and cellular immunity and hyper-IgE. The advent of next-generation sequencing technologies has enabled the rapid molecular diagnosis of rare monogenic diseases, including inborn errors of immunity. These advances have resulted in the implementation of gene-guided treatments, such as hematopoietic stem cell transplant for DOCK8 deficiency. However, putative disease-causing variants revealed by next-generation sequencing need rigorous validation to demonstrate pathogenicity. Here, we report the eventual diagnosis of DOCK8 deficiency in a consanguineous family due to a novel homozygous intronic deletion variant that caused aberrant exon splicing and subsequent loss of expression of DOCK8 protein. Remarkably, the causative variant was not initially detected by clinical whole-genome sequencing but was subsequently identified and validated by combining advanced genomic analysis, RNA-seq, and flow cytometry. This case highlights the need to adopt multipronged confirmatory approaches to definitively solve complex genetic cases that result from variants outside protein-coding exons and conventional splice sites.



Similar content being viewed by others
Availability of Data and Materials
Available upon request to the corresponding author.
Code Availability
Not applicable.
References
Tangye SG, Bucciol G, Casas-Martin J, Pillay B, Ma CS, Moens L, et al. Human inborn errors of the actin cytoskeleton affecting immunity: way beyond WAS and WIP. Immunol Cell Biol. 2019;97(4):389–402. https://doi.org/10.1111/imcb.12243.
Chen Y, Chen Y, Yin W, Han H, Miller H, Li J, et al. The regulation of DOCK family proteins on T and B cells. J Leukoc Biol. 2021;109(2):383–94. https://doi.org/10.1002/JLB.1MR0520-221RR.
Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361(21):2046–55. https://doi.org/10.1056/NEJMoa0905506.
Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124(6):1289-302 e4. https://doi.org/10.1016/j.jaci.2009.10.038.
Su HC, Jing H, Angelus P, Freeman AF. Insights into immunity from clinical and basic science studies of DOCK8 immunodeficiency syndrome. Immunol Rev. 2019;287(1):9–19. https://doi.org/10.1111/imr.12723.
Zhang Q, Davis JC, Dove CG, Su HC. Genetic, clinical, and laboratory markers for DOCK8 immunodeficiency syndrome. Dis Markers. 2010;29(3–4):131–9. https://doi.org/10.3233/DMA-2010-0737.
Aydin SE, Kilic SS, Aytekin C, Kumar A, Porras O, Kainulainen L, et al. DOCK8 deficiency: clinical and immunological phenotype and treatment options — a review of 136 patients. J Clin Immunol. 2015;35(2):189–98. https://doi.org/10.1007/s10875-014-0126-0.
Engelhardt KR, Gertz ME, Keles S, Schaffer AA, Sigmund EC, Glocker C, et al. The extended clinical phenotype of 64 patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol. 2015;136(2):402–12. https://doi.org/10.1016/j.jaci.2014.12.1945.
Al-Herz W, Chu JI, van der Spek J, Raghupathy R, Massaad MJ, Keles S, et al. Hematopoietic stem cell transplantation outcomes for 11 patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol. 2016;138(3):852-9 e3. https://doi.org/10.1016/j.jaci.2016.02.022.
Aydin SE, Freeman AF, Al-Herz W, Al-Mousa HA, Arnaout RK, Aydin RC, et al. Hematopoietic stem cell transplantation as treatment for patients with DOCK8 deficiency. J Allergy Clin Immunol Pract. 2019;7(3):848–55. https://doi.org/10.1016/j.jaip.2018.10.035.
Pillay BA, Avery DT, Smart JM, Cole T, Choo S, Chan D, et al. Hematopoietic stem cell transplant effectively rescues lymphocyte differentiation and function in DOCK8-deficient patients. JCI Insight. 2019;5. https://doi.org/10.1172/jci.insight.127527.
Haskologlu S, Kostel Bal S, Islamoglu C, Aytekin C, Guner S, Sevinc S, et al. Clinical, immunological features and follow up of 20 patients with dedicator of cytokinesis 8 (DOCK8) deficiency. Pediatr Allergy Immunol. 2020;31(5):515–27. https://doi.org/10.1111/pai.13236.
Chinn IK, Chan AY, Chen K, Chou J, Dorsey MJ, Hajjar J, et al. Diagnostic interpretation of genetic studies in patients with primary immunodeficiency diseases: a working group report of the Primary Immunodeficiency Diseases Committee of the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2020;145(1):46–69. https://doi.org/10.1016/j.jaci.2019.09.009.
Pillay BA, Fusaro M, Gray PE, Statham AL, Burnett L, Bezrodnik L, et al. Somatic reversion of pathogenic DOCK8 variants alters lymphocyte differentiation and function to effectively cure DOCK8 deficiency. J Clin Invest. 2021;131(3). https://doi.org/10.1172/JCI142434.
Randall KL, Chan SS, Ma CS, Fung I, Mei Y, Yabas M, et al. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J Exp Med. 2011;208(11):2305–20. https://doi.org/10.1084/jem.20110345.
Avery DT, Kane A, Nguyen T, Lau A, Nguyen A, Lenthall H, et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J Exp Med. 2018;215(8):2073–95. https://doi.org/10.1084/jem.20180010.
Pai SY, de Boer H, Massaad MJ, Chatila TA, Keles S, Jabara HH, et al. Flow cytometry diagnosis of dedicator of cytokinesis 8 (DOCK8) deficiency. J Allergy Clin Immunol. 2014;134(1):221–3. https://doi.org/10.1016/j.jaci.2014.02.023.
Tangye SG, Pillay B, Randall KL, Avery DT, Phan TG, Gray P, et al. Dedicator of cytokinesis 8-deficient CD4(+) T cells are biased to a TH2 effector fate at the expense of TH1 and TH17 cells. J Allergy Clin Immunol. 2017;139(3):933–49. https://doi.org/10.1016/j.jaci.2016.07.016.
Matthijs G, Souche E, Alders M, Corveleyn A, Eck S, Feenstra I, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24(1):2–5. https://doi.org/10.1038/ejhg.2015.226.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15. https://doi.org/10.1038/s41587-019-0201-4.
Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–73. https://doi.org/10.1093/nar/gky955.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. https://doi.org/10.1038/nbt.1754.
Brennan P. drawProteins: a Bioconductor/R package for reproducible and programmatic generation of protein schematics. F1000Res. 2018;7:1105. https://doi.org/10.12688/f1000research.14541.1.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021. https://doi.org/10.1038/s41586-021-03819-2.
Ma CS, Tangye SG. Flow cytometric-based analysis of defects in lymphocyte differentiation and function due to inborn errors of immunity. Front Immunol. 2019;10:2108. https://doi.org/10.3389/fimmu.2019.02108.
Crequer A, Troeger A, Patin E, Ma CS, Picard C, Pedergnana V, et al. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J Clin Invest. 2012;122(9):3239–47. https://doi.org/10.1172/JCI62949.
Moran I, Nguyen A, Khoo WH, Butt D, Bourne K, Young C, et al. Memory B cells are reactivated in subcapsular proliferative foci of lymph nodes. Nat Commun. 2018;9(1):3372. https://doi.org/10.1038/s41467-018-05772-7.
Bucciol G, Pillay B, Casas-Martin J, Delafontaine S, Proesmans M, Lorent N, et al. Systemic Inflammation and myelofibrosis in a patient with Takenouchi-Kosaki Syndrome due to CDC42 Tyr64Cys mutation. J Clin Immunol. 2020;40(4):567–70. https://doi.org/10.1007/s10875-020-00742-5.
Edwards ESJ, Bier J, Cole TS, Wong M, Hsu P, Berglund LJ, et al. Activating PIK3CD mutations impair human cytotoxic lymphocyte differentiation and function and EBV immunity. J Allergy Clin Immunol. 2019;143(1):276-91 e6. https://doi.org/10.1016/j.jaci.2018.04.030.
Jing H, Zhang Q, Zhang Y, Hill BJ, Dove CG, Gelfand EW, et al. Somatic reversion in dedicator of cytokinesis 8 immunodeficiency modulates disease phenotype. J Allergy Clin Immunol. 2014;133(6):1667–75. https://doi.org/10.1016/j.jaci.2014.03.025.
Picard C, Bobby Gaspar H, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J Clin Immunol. 2018;38(1):96–128. https://doi.org/10.1007/s10875-017-0464-9.
Phan TG, Gray PE, Wong M, Macintosh R, Burnett L, Tangye SG, et al. The Clinical Immunogenomics Research Consortium Australasia (CIRCA): a distributed network model for genomic healthcare delivery. J Clin Immunol. 2020;40(5):763–6. https://doi.org/10.1007/s10875-020-00787-6.
Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. Human inborn errors of immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2020;40(1):24–64. https://doi.org/10.1007/s10875-019-00737-x.
Minoche AE, Lundie B, Peters GB, Ohnesorg T, Pinese M, Thomas DM, et al. ClinSV: clinical grade structural and copy number variant detection from whole genome sequencing data. Genome Med. 2021;13(1):32. https://doi.org/10.1186/s13073-021-00841-x.
Ito K, Patel PN, Gorham JM, McDonough B, DePalma SR, Adler EE, et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc Natl Acad Sci U S A. 2017;114(29):7689–94. https://doi.org/10.1073/pnas.1707741114.
Ribeiro M, Furtado M, Martins S, Carvalho T, Carmo-Fonseca M. RNA splicing defects in hypertrophic cardiomyopathy: implications for diagnosis and therapy. Int J Mol Sci. 2020;21(4):1329. https://doi.org/10.3390/ijms21041329.
Xu X, Han L, Zhao G, Xue S, Gao Y, Xiao J, et al. LRCH1 interferes with DOCK8-Cdc42-induced T cell migration and ameliorates experimental autoimmune encephalomyelitis. J Exp Med. 2017;214(1):209–26. https://doi.org/10.1084/jem.20160068.
Meyts I, Bosch B, Bolze A, Boisson B, Itan Y, Belkadi A, et al. Exome and genome sequencing for inborn errors of immunity. J Allergy Clin Immunol. 2016;138(4):957–69. https://doi.org/10.1016/j.jaci.2016.08.003.
Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. The ever-increasing array of novel inborn errors of immunity: an interim update by the IUIS Committee. J Clin Immunol. 2021;41(3):666–79. https://doi.org/10.1007/s10875-021-00980-1.
Stray-Pedersen A, Sorte HS, Samarakoon P, Gambin T, Chinn IK, Coban Akdemir ZH, et al. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol. 2017;139(1):232–45. https://doi.org/10.1016/j.jaci.2016.05.042.
Sullivan KE. The scary world of variants of uncertain significance (VUS): a hitchhiker’s guide to interpretation. J Allergy Clin Immunol. 2021;147(2):492–4. https://doi.org/10.1016/j.jaci.2020.06.011.
Weck KE. Interpretation of genomic sequencing: variants should be considered uncertain until proven guilty. Genet Med. 2018;20(3):291–3. https://doi.org/10.1038/gim.2017.269.
Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9(1):26. https://doi.org/10.1186/s13073-017-0412-6.
Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. https://doi.org/10.1186/1741-7007-7-23.
Khourieh J, Rao G, Habib T, Avery DT, Lefevre-Utile A, Chandesris MO, et al. A deep intronic splice mutation of STAT3 underlies hyper IgE syndrome by negative dominance. Proc Natl Acad Sci U S A. 2019;116(33):16463–72. https://doi.org/10.1073/pnas.1901409116.
Boisson B, Honda Y, Ajiro M, Bustamante J, Bendavid M, Gennery AR, et al. Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency. J Clin Invest. 2019;129(2):583–97. https://doi.org/10.1172/JCI124011.
Khan S, Kuruvilla M, Hagin D, Wakeland B, Liang C, Vishwanathan K, et al. RNA sequencing reveals the consequences of a novel insertion in dedicator of cytokinesis-8. J Allergy Clin Immunol. 2016;138(1):289-92 e6. https://doi.org/10.1016/j.jaci.2015.11.033.
Acknowledgements
We are extremely thankful to the patients and their families for participating in this study. We also thank Angela Wang (NIAID, NIH) for her regulatory assistance; the Clinical Trials & Biorepository Team, St Vincent’s Centre for Applied Medical Research (St Vincent’s Hospital, Darlinghurst, NSW Australia), for biobanking; and Dr. Karen Enthoven (CIRCA) for project coordination. Computational work was performed using the high-performance computing resources of the Garvan Institute of Medical Research. Sanger sequencing was performed by Garvan Molecular Genetics and flow cytometry by the Garvan Flow facility.
Funding
This study was supported by the NHMRC of Australia (1060303), Office of Health and Medical Research of the NSW Government, the Jeffrey Modell Foundation, SPHERE Triple I Clinical Academic Group and UNSW Medicine Infection, Immunology and Inflammation Theme, the Ross Trust, and the John Brown Cook Foundation, and in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH. S.G.T was a Principal Research Fellow (1042925) of the NHMRC and is currently a recipient of an NHMRC Leadership 3 Investigator Grant (1176665) and NHMRC program grant (1113904). C.S.M is supported by an Early-Mid Career Research Fellowship from the Ministry of Health of the New South Wales Government of Australia.
Author information
Authors and Affiliations
Contributions
SGT, PG, LB, and CSM conceived and designed the study; PG, BP, JYY, JR, WAF, SK, and CSM conducted experiments; DEC, AS, and JP provided patient care and data collection; JS and GU performed Ion Torrent, and HJ and HCS performed initial DOCK8 exon sequencing; SGT, PG, LB, and CSM supervised the study; SGT and CSM wrote drafts of the original and revised manuscripts; all authors contributed to the final version of the manuscript and approved submission of the final version.
Corresponding author
Ethics declarations
Ethics Approval
This study was approved by the Sydney Local Health District RPAH Zone Human Research Ethics Committee and Research Governance Office, Royal Prince Alfred Hospital, Camperdown, NSW, Australia (Protocol X16-0210/LNR/16/RPAH/257); the South East Sydney Local Health District Human Research Ethics Committee, Prince of Wales/Sydney Children’s Hospital, Randwick, NSW, Australia (Protocol HREC/11/POWH/152); and NIAID (Protocol 06-I-0015). Written informed consent was obtained from participants or their guardians.
Consent to Participate
Written informed consent was obtained from participants or their guardians.
Consent for Publication
Patients signed informed consent regarding publishing their data.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tangye, S.G., Gray, P.E., Pillay, B.A. et al. Hyper-IgE Syndrome due to an Elusive Novel Intronic Homozygous Variant in DOCK8. J Clin Immunol 42, 119–129 (2022). https://doi.org/10.1007/s10875-021-01152-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-021-01152-x