Abstract
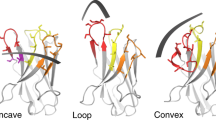
The insertase BamA is an essential protein of the bacterial outer membrane. Its 16-stranded transmembrane β-barrel contains a lateral gate as a key functional element. This gate is formed by the C-terminal half of the last β-strand. The BamA barrel was previously found to sample different conformations in aqueous solution, as well as different gate-open, gate-closed, and collapsed conformations in X-ray crystallography and cryo-electron microscopy structures. Here, we report the successful identification of conformation-selective nanobodies that stabilize BamA in specific conformations. While the initial candidate generation and selection protocol was based on established alpaca immunization and phage display selection procedures, the final selection of nanobodies was enhanced by a solution NMR-based screening step to shortlist the targets for crystallization. In this way, three crystal structures of BamA–nanobody complexes were efficiently obtained, showing two types of nanobodies that indeed stabilized BamA in two different conformations, i.e., with open and closed lateral gate, respectively. Then, by correlating the structural data with high resolution NMR spectra, we could for the first time assign the BamA conformational solution ensemble to defined structural states. The new nanobodies will be valuable tools towards understanding the client insertion mechanism of BamA and towards developing improved antibiotics.





Similar content being viewed by others
References
Adams PD et al (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954
Albrecht R et al (2014) Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr 70:1779–1789
Bakelar J, Buchanan SK, Noinaj N (2016) The structure of the β-barrel assembly machinery complex. Science 351:180–186
Blanpain C et al (2002) Multiple active states and oligomerization of CCR25 revealed by functional properties of monoclonal antibodies. Mol Biol Cell 13:723–737
Chen VB et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21
De Geyter J et al (2016) Protein folding in the cell envelope of Escherichia coli. Nat Microbiol 1:16107
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40:82–92
Dmitriev OY, Lutsenko S, Muyldermans S (2016) Nanobodies as probes for protein dynamics in vitro and in cells. J Biol Chem 291:3767–3775
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Evans PR, Murshudov GN (2013) How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214
Garcia-Nafria J, Lee Y, Bai X, Carpenter B, Tate CG (2018) Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Elife 7:e35946
Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164:19–24
Gu Y et al (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69
Hagan CL, Silhavy TJ, Kahne D (2011) β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem 80:189–210
Hamers-Casterman C et al (1993) Naturally occurring antibodies devoid of light chains. Nature 363:446–448
Han L et al (2016) Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196
Hartmann JB, Zahn M, Burmann IM, Bibow S, Hiller S (2018) Sequence-specific solution NMR assignments of the β-Barrel Insertase BamA to monitor its conformational ensemble at the atomic level. J Am Chem Soc 140:11252–11260
Iadanza MG et al (2016) Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865
Ji X et al (2017) Neutralization of TNFalpha in tumor with a novel nanobody potentiates paclitaxel-therapy and inhibits metastasis in breast cancer. Cancer Lett 386:24–34
Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66:125–132
Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7:206–214
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Kumar H et al (2018) Crystal structure of a ligand-bound LacY–nanobody complex. Proc Natl Acad Sci U S A 115:8769–8774
Masureel M et al (2018) Structural insights into binding specificity, efficacy and bias of a β2AR partial agonist. Nat Chem Biol 14:1059–1066
Mireku SA, Sauer MM, Glockshuber R, Locher KP (2017) Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD. Sci Rep 7:14296
Murshudov GN et al (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367
Ni D et al (2014) Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 28:2677–2685
Pardon E et al (2014) A general protocol for the generation of nanobodies for structural biology. Nat Protoc 9:674–693
Perez C et al (2017) Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody. Sci Rep 7:46641
Pinto C et al (2018) Formation of the β-barrel assembly machinery complex in lipid bilayers as seen by solid-state NMR. Nat Commun 9:4135
Rasmussen SG et al (2011a) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469:175–180
Rasmussen SG et al (2011b) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477:549–555
Ricci DP, Silhavy TJ (2012) The Bam machine: a molecular cooper. Biochim Biophys Acta 1818:1067–1084
Ricci DP, Hagan CL, Kahne D, Silhavy TJ (2012) Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109:3487–3491
Rigel NW, Schwalm J, Ricci DP, Silhavy TJ (2012) BamE modulates the Escherichia coli β-barrel assembly machine component BamA. J Bacteriol 194:1002–1008
Storek KM et al (2018) Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A 115:3692–3697
Terwilliger TC et al (2008) Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr D Biol Crystallogr 64:515–524
Vagin A, Teplyakov A (2010) Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66:22–25
Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265
Zavrtanik U, Lukan J, Loris R, Lah J, Hadzi S (2018) Structural basis of epitope recognition by heavy-chain camelid antibodies. J Mol Biol 430:4369–4386
Zimmermann I et al (2018) Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife 7:e34317
Acknowledgements
We thank Tim Sharpe for scientific support and discussions and sciCORE at University of Basel for support with high performance computing. S. Štefanić from the Nanobody Service Facility of the University of Zurich is acknowledged for alpaca immunizations. X-ray diffraction data were collected at beamline PXI of the Paul Scherrer Institute, Villigen, Switzerland; we acknowledge excellent support by the facility team. Funding from the Swiss National Science Foundation via the NFP 72 (407240_167125) to S.H., by an SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823) to M.A.S. and by a BioEntrepreneur-Fellowship of the University of Zurich (BIOEF-17-002) to I.Z. is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaur, H., Hartmann, JB., Jakob, R.P. et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J Biomol NMR 73, 375–384 (2019). https://doi.org/10.1007/s10858-019-00250-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-019-00250-8